5MCO
 
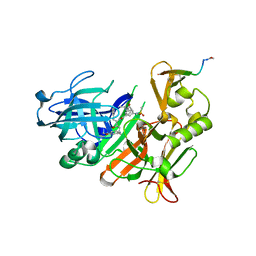 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE INHIBITOR GRL-8234 AND EXOSITE PEPTIDE | | Descriptor: | BACE-1 EXOSITE PEPTIDE, Beta-secretase 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5OR0
 
 | |
5SAK
 
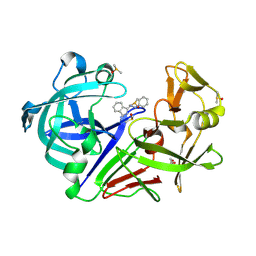 | | Endothiapepsin in complex with compound FU5-1 | | Descriptor: | (1Z,3Z)-3-(2-phenylhydrazinylidene)-2,3-dihydro-1H-isoindol-1-imine, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAQ
 
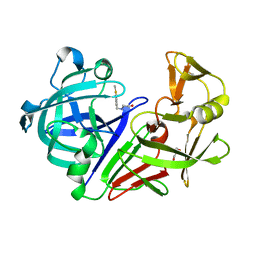 | | Endothiapepsin in complex with compound FU58-3 | | Descriptor: | 2-({[4-(methylsulfanyl)phenyl]methyl}amino)ethan-1-ol, Endothiapepsin, GLYCEROL | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAR
 
 | | Endothiapepsin in complex with compound FU290-1 | | Descriptor: | (1,4-phenylene)bis(methylene) dicarbamimidothioate, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAO
 
 | | Endothiapepsin in complex with compound FU58-1 | | Descriptor: | 1,2-ETHANEDIOL, 6-[(8R)-2-({[(3,5-dimethyl-1,2-oxazol-4-yl)methyl](methyl)amino}methyl)-6,7-dihydropyrazolo[1,5-a]pyrazin-5(4H)-yl]pyrimidin-4-amine, Endothiapepsin, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAL
 
 | | Endothiapepsin in complex with compound FU5-2 | | Descriptor: | (1Z)-1-imino-1H-isoindol-3-amine, DIMETHYL SULFOXIDE, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAT
 
 | | Endothiapepsin in complex with compound FU66-1 | | Descriptor: | Endothiapepsin, GLYCEROL, N-cyclopropyl-6-(furan-2-yl)-2-hydroxy-N-[(pyridin-2-yl)methyl]pyridine-3-carboxamide | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAN
 
 | | Endothiapepsin in complex with compound FU5-4 | | Descriptor: | (2Z)-2-(3-amino-1H-isoindol-1-ylidene)hydrazine-1-carboximidamide, Endothiapepsin, GLYCEROL | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAM
 
 | | Endothiapepsin in complex with compound FU5-3 | | Descriptor: | 3-amino-1H-isoindol-1-one, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAS
 
 | | Endothiapepsin in complex with compound FU290-2 | | Descriptor: | (1R)-1-(4-chlorophenyl)ethyl carbamimidothioate, BROMIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5SAP
 
 | | Endothiapepsin in complex with compound FU58-2 | | Descriptor: | (8S)-5-(2-aminopyrimidin-4-yl)-N-[2-(dimethylamino)ethyl]-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazine-2-carboxamide, 1,2-ETHANEDIOL, Endothiapepsin, ... | | Authors: | Wollenhaupt, J, Metz, A, Messini, N, Barthel, T, Klebe, G, Weiss, M.S. | | Deposit date: | 2021-05-28 | | Release date: | 2021-09-01 | | Last modified: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Frag4Lead: growing crystallographic fragment hits by catalog using fragment-guided template docking.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5QU8
 
 | |
5QU3
 
 | |
5QUA
 
 | | Crystal Structure of swapped human Nck SH3.1 domain, 1.5A, C2221 | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU1
 
 | | Crystal Structure of the monomeric human Nck SH3.1 domain, triclinic, 1.08A | | Descriptor: | Cytoplasmic protein NCK1, SULFATE ION | | Authors: | Burger, D, Ruf, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU5
 
 | | Domain Swap in the first SH3 domain of human Nck1 | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Burger, D, Ruf, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU2
 
 | | Crystal Structure of human Nck SH3.1 in complex with peptide PPPVPNPDY | | Descriptor: | ACE-PRO-PRO-PRO-VAL-PRO-ASN-PRO-ASP-TYR-NH2, Cytoplasmic protein NCK1, SULFATE ION | | Authors: | Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
5QU7
 
 | |
5QU4
 
 | |
5QU6
 
 | | Crystal Structure of swapped human Nck SH3.1 domain, 1.8A, triclinic | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Rudolph, M.G. | | Deposit date: | 2019-12-13 | | Release date: | 2020-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Small molecule AX-024 reduces T cell proliferation independently of CD3ε/Nck1 interaction, which is governed by a domain swap in the Nck1-SH3.1 domain.
J.Biol.Chem., 295, 2020
|
|
1KXO
 
 | | ENGINEERED LIPOCALIN DIGA16 : APO-FORM | | Descriptor: | DigA16 | | Authors: | Korndoerfer, I.P, Skerra, A. | | Deposit date: | 2002-02-01 | | Release date: | 2003-06-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural mechanism of specific ligand recognition by a lipocalin tailored for the complexation of digoxigenin.
J.Mol.Biol., 330, 2003
|
|
1LNM
 
 | |
1LKE
 
 | |
3BX8
 
 | | Engineered Human Lipocalin 2 (LCN2), apo-form | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ENGINEERED HUMAN LIPOCALIN 2, PENTAETHYLENE GLYCOL | | Authors: | Schonfeld, D.L, Chatwell, L, Skerra, A. | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High affinity molecular recognition and functional blockade of CTLA-4 by an engineered human lipocalin
To be Published
|
|
