6MR5
 
 | |
6NKE
 
 | | Wild-type GGGPS from Thermoplasma volcanium | | Descriptor: | GLYCEROL, Geranylgeranylglyceryl phosphate synthase, IMIDAZOLE, ... | | Authors: | Blank, P.N, Alderfer, K.E, Gillott, B.N, Christianson, D.W, Himmelberger, J.A. | | Deposit date: | 2019-01-07 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural studies of geranylgeranylglyceryl phosphate synthase, a prenyltransferase found in thermophilic Euryarchaeota.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6O9P
 
 | | Wild-type SaSQS1 Complexed with Ibandronate | | Descriptor: | 1,2-ETHANEDIOL, IBANDRONATE, MAGNESIUM ION, ... | | Authors: | Blank, P.N, Christianson, D.W. | | Deposit date: | 2019-03-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Sesquisabinene Synthase 1, a Terpenoid Cyclase That Generates a Strained [3.1.0] Bridged-Bicyclic Product.
Acs Chem.Biol., 14, 2019
|
|
6O9Q
 
 | | Wild-type SaSQS1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Sesquisabinene B synthase 1 | | Authors: | Blank, P.N, Christianson, D.W. | | Deposit date: | 2019-03-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Sesquisabinene Synthase 1, a Terpenoid Cyclase That Generates a Strained [3.1.0] Bridged-Bicyclic Product.
Acs Chem.Biol., 14, 2019
|
|
8DAY
 
 | | Crystal Structure of DMATS1 prenyltransferase in complex with L-Tyr and DMSPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, Dimethylallyltryptophan synthase 1, TYROSINE | | Authors: | Eaton, S.A, Ronnebaum, T.A, Roose, B.W, Christianson, D.W. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Substrate Promiscuity and Catalysis by the Reverse Prenyltransferase N -Dimethylallyl-l-tryptophan Synthase from Fusarium fujikuroi .
Biochemistry, 61, 2022
|
|
8DAZ
 
 | | Crystal structure of DMATS1 prenyltransferase in complex with L-Trp and GSPP | | Descriptor: | Dimethylallyltryptophan synthase 1, GERANYL S-THIOLODIPHOSPHATE, TRYPTOPHAN | | Authors: | Eaton, S.A, Ronnebaum, T.A, Roose, B.W, Christianson, D.W. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Basis of Substrate Promiscuity and Catalysis by the Reverse Prenyltransferase N -Dimethylallyl-l-tryptophan Synthase from Fusarium fujikuroi .
Biochemistry, 61, 2022
|
|
8DB1
 
 | | Crystal structure of native DMATS1 prenyltransferase | | Descriptor: | Dimethylallyltryptophan synthase 1, TRYPTOPHAN | | Authors: | Eaton, S.A, Ronnebaum, T.A, Roose, B.W, Christianson, D.W. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Substrate Promiscuity and Catalysis by the Reverse Prenyltransferase N -Dimethylallyl-l-tryptophan Synthase from Fusarium fujikuroi .
Biochemistry, 61, 2022
|
|
8DB0
 
 | | Crystal structure of DMATS1 prenyltransferase in complex with L-Trp and DMSPP | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Eaton, S.A, Ronnebaum, T.A, Roose, B.W, Christianson, D.W. | | Deposit date: | 2022-06-14 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural Basis of Substrate Promiscuity and Catalysis by the Reverse Prenyltransferase N -Dimethylallyl-l-tryptophan Synthase from Fusarium fujikuroi .
Biochemistry, 61, 2022
|
|
4QA6
 
 | | Crystal structure of I243N/Y306F HDAC8 in complex with a tetrapeptide substrate | | Descriptor: | 7-AMINO-4-METHYL-CHROMEN-2-ONE, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
4QA2
 
 | | Crystal structure of I243N HDAC8 in complex with SAHA | | Descriptor: | GLYCEROL, Histone deacetylase 8, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.377 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
4QA1
 
 | | Crystal structure of A188T HDAC8 in complex with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
4QA4
 
 | | Crystal structure of H334R HDAC8 in complex with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
4QA7
 
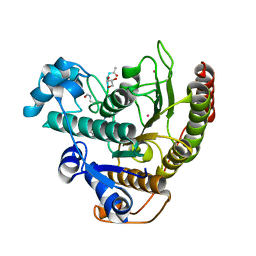 | | Crystal structure of H334R/Y306F HDAC8 in complex with a tetrapeptide substrate | | Descriptor: | 7-AMINO-4-METHYL-CHROMEN-2-ONE, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
4ZUO
 
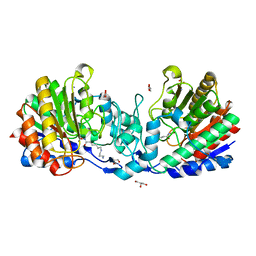 | | Crystal structure of acetylpolyamine amidohydrolase from Mycoplana ramosa in complex with a hydroxamate inhibitor | | Descriptor: | 6-[(3-aminopropyl)amino]-N-hydroxyhexanamide, AMMONIUM ION, Acetylpolyamine aminohydrolase, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2015-05-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase.
Biochemistry, 54, 2015
|
|
4ZUQ
 
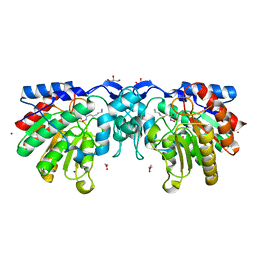 | | Crystal structure of acetylpolyamine amidohydrolase from Mycoplana ramosa in complex with a hydroxamate inhibitor | | Descriptor: | 6-amino-N-hydroxyhexanamide, Acetylpolyamine aminohydrolase, GLYCEROL, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2015-05-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase.
Biochemistry, 54, 2015
|
|
4ZUP
 
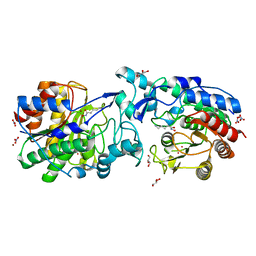 | | Crystal structure of acetylpolyamine amidohydrolase from Mycoplana ramosa in complex with a hydroxamate inhibitor | | Descriptor: | 5-amino-N-hydroxypentanamide, Acetylpolyamine aminohydrolase, GLYCEROL, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2015-05-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase.
Biochemistry, 54, 2015
|
|
4ZUN
 
 | | Crystal structure of acetylpolyamine amidohydrolase from Mycoplana ramosa in complex with a thiol inhibitor | | Descriptor: | 5-[(3-aminopropyl)amino]pentane-1-thiol, Acetylpolyamine aminohydrolase, GLYCEROL, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2015-05-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase.
Biochemistry, 54, 2015
|
|
4QA0
 
 | | Crystal structure of C153F HDAC8 in complex with SAHA | | Descriptor: | GLYCEROL, Histone deacetylase 8, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
4QA3
 
 | | Crystal structure of T311M HDAC8 in complex with Trichostatin A (TSA) | | Descriptor: | GLYCEROL, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.876 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
4ZUM
 
 | | Crystal structure of acetylpolyamine amidohydrolase from Mycoplana ramosa in complex with a trifluoromethylketone inhibitor | | Descriptor: | 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptane-2,2-diol, Acetylpolyamine aminohydrolase, GLYCEROL, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2015-05-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase.
Biochemistry, 54, 2015
|
|
4ZUR
 
 | | Crystal structure of acetylpolyamine amidohydrolase from Mycoplana ramosa in complex with a hydroxamate inhibitor | | Descriptor: | 7-amino-N-hydroxyheptanamide, AMMONIUM ION, Acetylpolyamine aminohydrolase, ... | | Authors: | Decroos, C, Christianson, D.W. | | Deposit date: | 2015-05-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Design, Synthesis, and Evaluation of Polyamine Deacetylase Inhibitors, and High-Resolution Crystal Structures of Their Complexes with Acetylpolyamine Amidohydrolase.
Biochemistry, 54, 2015
|
|
8EAX
 
 | |
8EQI
 
 | |
4QA5
 
 | | Crystal structure of A188T/Y306F HDAC8 in complex with a tetrapeptide substrate | | Descriptor: | 7-AMINO-4-METHYL-CHROMEN-2-ONE, GLYCEROL, Histone deacetylase 8, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
3G4D
 
 | | Crystal Structure of (+)-delta-Cadinene Synthase from Gossypium arboreum and Evolutionary Divergence of Metal Binding Motifs for Catalysis | | Descriptor: | (+)-delta-cadinene synthase isozyme XC1, BETA-MERCAPTOETHANOL, GLYCEROL | | Authors: | Gennadios, H.A, Di Costanzo, L, Miller, D.J, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2009-02-03 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structure of (+)-delta-cadinene synthase from Gossypium arboreum and evolutionary divergence of metal binding motifs for catalysis.
Biochemistry, 48, 2009
|
|
