3CP9
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyridone inhibitor | | Descriptor: | 3-(2-aminoquinazolin-6-yl)-1-(3,3-dimethylindolin-6-yl)-4-methylpyridin-2(1H)-one, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2008-03-31 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Aryl Aminoquinazoline Pyridones as Potent, Selective, and Orally Efficacious Inhibitors of Receptor Tyrosine Kinase c-Kit.
J.Med.Chem., 51, 2008
|
|
3DTW
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a benzisoxazole inhibitor | | Descriptor: | 6-chloro-N-pyrimidin-5-yl-3-{[3-(trifluoromethyl)phenyl]amino}-1,2-benzisoxazole-7-carboxamide, Vascular endothelial growth factor receptor 2 | | Authors: | Whittington, D.A, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | Deposit date: | 2008-07-16 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of amido-benzisoxazoles as potent c-Kit inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8WRZ
 
 | | Cry-EM structure of cannabinoid receptor-beta-arrestin-1 complex | | Descriptor: | (6~{a}~{R},9~{R},10~{a}~{R})-9-(hydroxymethyl)-3-(8-isothiocyanato-2-methyl-octan-2-yl)-6,6-dimethyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, Beta-arrestin-1, Soluble cytochrome b562,Cannabinoid receptor 1, ... | | Authors: | Wang, Y.X, Wang, T, Wu, L.J, Hua, T, Liu, Z.J. | | Deposit date: | 2023-10-16 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of cannabinoid receptor CB1-beta-arrestin complex.
Protein Cell, 15, 2024
|
|
7Y3G
 
 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | G-protein coupled receptor 12, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Li, H, Zhang, J.Y, Luo, F. | | Deposit date: | 2022-06-10 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insight into the constitutive activity of human orphan receptor GPR12.
Sci Bull (Beijing), 68, 2023
|
|
4DDL
 
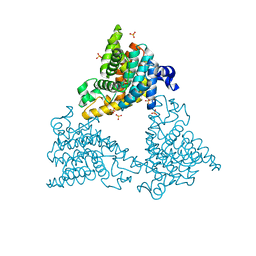 | | PDE10a Crystal Structure Complexed with Novel Inhibitor | | Descriptor: | 2-{1-[5-(6,7-dimethoxycinnolin-4-yl)-3-methylpyridin-2-yl]piperidin-4-yl}propan-2-ol, SULFATE ION, ZINC ION, ... | | Authors: | Chmait, S, Jordan, S, Zhang, J. | | Deposit date: | 2012-01-18 | | Release date: | 2012-03-21 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of potent, selective, and metabolically stable 4-(pyridin-3-yl)cinnolines as novel phosphodiesterase 10A (PDE10A) inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4HF4
 
 | |
4HEU
 
 | |
7VUG
 
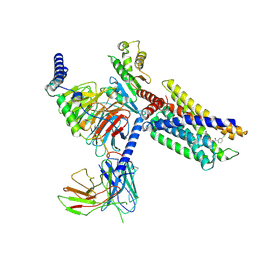 | | Cryo-EM structure of a class A orphan GPCR in complex with Gi | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Chimera of Endo-1,4-beta-xylanase and Probable G-protein coupled receptor 139, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUH
 
 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUJ
 
 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
7VUI
 
 | | Cryo-EM structure of a class A orphan GPCR | | Descriptor: | 3-chloranyl-N-[2-oxidanylidene-2-[[(1S)-1-phenylethyl]amino]ethyl]benzamide, Chimera of Endo-1,4-beta-xylanase and Probable G-protein coupled receptor 139, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Liu, Z.J, Hua, T, Zhou, Y.L, Wu, L.J. | | Deposit date: | 2021-11-02 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular insights into ligand recognition and G protein coupling of the neuromodulatory orphan receptor GPR139.
Cell Res., 32, 2022
|
|
6HR2
 
 | | Crystal structure of PROTAC 2 in complex with the bromodomain of human SMARCA4 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[4-[[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]methyl]phenyl]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
6FR2
 
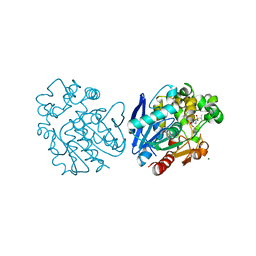 | | Soluble epoxide hydrolase in complex with LK864 | | Descriptor: | 1-[(4~{S})-9-propan-2-ylsulfonyl-1-oxa-9-azaspiro[5.5]undecan-4-yl]-3-[[4-(trifluoromethyloxy)phenyl]methyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION | | Authors: | Kramer, J.S, Pogoryelov, D, Krasavin, M, Proschak, E. | | Deposit date: | 2018-02-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Discovery of polar spirocyclic orally bioavailable urea inhibitors of soluble epoxide hydrolase.
Bioorg. Chem., 80, 2018
|
|
4WUU
 
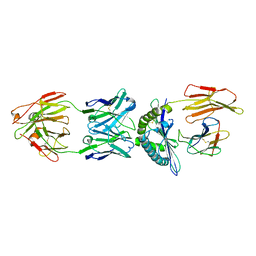 | | Structure of ESK1 in complex with HLA-A*0201/WT1 | | Descriptor: | ARG-MET-PHE-PRO-ASN-ALA-PRO-TYR-LEU, Beta-2-microglobulin, ESK1, ... | | Authors: | Ataie, N.J, Ng, H.L. | | Deposit date: | 2014-11-03 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.047 Å) | | Cite: | Structure of a TCR-Mimic Antibody with Target Predicts Pharmacogenetics.
J.Mol.Biol., 428, 2016
|
|
5DHV
 
 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
5DHZ
 
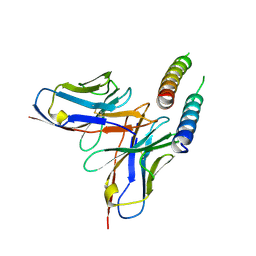 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
5DHX
 
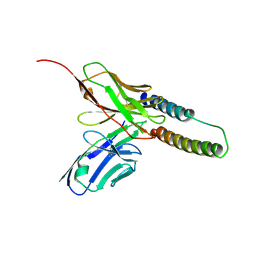 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, light chain,Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
5DHY
 
 | | HIV-1 Rev NTD dimers with variable crossing angles | | Descriptor: | Anti-Rev Antibody Fab single-chain variable fragment, heavy chain, light chain, ... | | Authors: | DiMattia, M.A, Watts, N.R, Wingfield, P.T, Grimes, J.M, Stuart, D.I, Steven, A.C. | | Deposit date: | 2015-08-31 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Structure of HIV-1 Rev Filaments Suggests a Bilateral Model for Rev-RRE Assembly.
Structure, 24, 2016
|
|
5EQ0
 
 | | Crystal Structure of chromodomain of CBX8 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 8, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-12 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
5EPK
 
 | | Crystal Structure of chromodomain of CBX2 in complex with inhibitor UNC3866 | | Descriptor: | Chromobox protein homolog 2, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
5EPL
 
 | | Crystal Structure of chromodomain of CBX4 in complex with inhibitor UNC3866 | | Descriptor: | E3 SUMO-protein ligase CBX4, UNKNOWN ATOM OR ION, unc3866 | | Authors: | Liu, Y, Tempel, W, Walker, J.R, Stuckey, J.I, Dickson, B.M, James, L.I, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-11 | | Release date: | 2015-12-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1.
Nat.Chem.Biol., 12, 2016
|
|
1SET
 
 | |
1SER
 
 | | THE 2.9 ANGSTROMS CRYSTAL STRUCTURE OF T. THERMOPHILUS SERYL-TRNA SYNTHETASE COMPLEXED WITH TRNA SER | | Descriptor: | PROTEIN (SERYL-TRNA SYNTHETASE (E.C.6.1.1.11)), TRNASER | | Authors: | Biou, S, Cusack, V, Yaremchuk, A, Tukalo, M. | | Deposit date: | 1994-02-21 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser).
Science, 263, 1994
|
|
1SES
 
 | |
1C5E
 
 | | BACTERIOPHAGE LAMBDA HEAD PROTEIN D | | Descriptor: | GLYCEROL, HEAD DECORATION PROTEIN | | Authors: | Yang, F, Forrer, P, Dauter, Z, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1999-11-18 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD.
Nat.Struct.Biol., 7, 2000
|
|
