2HY9
 
 | | Human telomere DNA quadruplex structure in K+ solution hybrid-1 form | | Descriptor: | DNA (26-MER) | | Authors: | Dai, J, Punchihewa, C, Jones, R.A, Hurley, L, Yang, D. | | Deposit date: | 2006-08-04 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple formation.
Nucleic Acids Res., 35, 2007
|
|
5JSG
 
 | | Crystal structure of Spindlin1 bound to compound EML405 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Spindlin-1, ... | | Authors: | Su, X, Li, H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Developing Spindlin1 small-molecule inhibitors by using protein microarrays
Nat. Chem. Biol., 13, 2017
|
|
5JSJ
 
 | | Crystal structure of Spindlin1 bound to compound EML631 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Spindlin-1, ... | | Authors: | Su, X, Li, H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.348 Å) | | Cite: | Developing Spindlin1 small-molecule inhibitors by using protein microarrays
Nat. Chem. Biol., 13, 2017
|
|
3DPK
 
 | | cFMS tyrosine kinase in complex with a pyridopyrimidinone inhibitor | | Descriptor: | 8-cyclohexyl-N-methoxy-5-oxo-2-{[4-(2-pyrrolidin-1-ylethyl)phenyl]amino}-5,8-dihydropyrido[2,3-d]pyrimidine-6-carboxamide, Macrophage colony-stimulating factor 1 receptor, Fibroblast growth factor receptor 1, ... | | Authors: | Schubert, C. | | Deposit date: | 2008-07-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrido[2,3-d]pyrimidin-5-ones: a novel class of antiinflammatory macrophage colony-stimulating factor-1 receptor inhibitors
J.Med.Chem., 52, 2009
|
|
8EKD
 
 | |
4NT4
 
 | | Crystal structure of the kinase domain of Gilgamesh isoform I from Drosophila melanogaster | | Descriptor: | GLYCEROL, Gilgamesh, isoform I, ... | | Authors: | Chen, C.C, Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2013-11-30 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of the kinase domain of Gilgamesh from Drosophila melanogaster
Acta Crystallogr.,Sect.F, 70, 2014
|
|
6F57
 
 | |
8H08
 
 | |
8H07
 
 | |
7ONP
 
 | | Wild type carbonic anhydrase II with bound IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 4-[2-(9-chloranyl-2',3',4',5',6'-pentamethyl-4-nitro-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, ACETATE ION, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.408 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7ONV
 
 | | Carbonic anhydrase II mutant (I91C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-26 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7ONM
 
 | | Carbonic anhydrase II mutant (N67G-E69R-I91C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7ONQ
 
 | | Carbonic anhydrase II mutant (E69C) dually binding an IrCp* complex to generate an artificial transfer hydrogenase (ATHase) | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-(4-azanyl-9-chloranyl-2',3',4',5',6'-pentamethyl-7-oxidanylidene-spiro[1$l^{4},8-diaza-9$l^{8}-iridabicyclo[4.3.0]nona-1,3,5-triene-9,1'-1$l^{8}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexane]-8-yl)ethyl]benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Stein, A, Dongping, C, Cotelle, Y, Rebelein, J.G, Ward, T.R. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Dual Anchoring Strategy for the Directed Evolution of Improved Artificial Transfer Hydrogenases Based on Carbonic Anhydrase.
Acs Cent.Sci., 7, 2021
|
|
7RAM
 
 | | Cryo-EM Structure of the HCMV gHgLgO Trimer Derived from AD169 and TR strains in complex with PDGFRalpha | | Descriptor: | Envelope glycoprotein H, Envelope glycoprotein L, Envelope glycoprotein O, ... | | Authors: | Liu, J, Vanarsdall, A.L, Chen, D, Johnson, D.C, Jardetzky, T.S. | | Deposit date: | 2021-07-02 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Cryo-Electron Microscopy Structure and Interactions of the Human Cytomegalovirus gHgLgO Trimer with Platelet-Derived Growth Factor Receptor Alpha.
Mbio, 12, 2021
|
|
5XGV
 
 | |
7RYX
 
 | | S. CEREVISIAE CYP51 COMPLEXED WITH VT-1129 | | Descriptor: | (2R)-2-(2,4-difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-{5-[4-(trifluoromethoxy)phenyl]pyridin-2-yl}propan-2-ol, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ruma, Y.N, Sagatova, A, Keniya, M.V, Tyndall, J.D, Monk, B.C. | | Deposit date: | 2021-08-26 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterisation of Candida parapsilosis CYP51 as a Drug Target Using Saccharomyces cerevisiae as Host.
J Fungi, 8, 2022
|
|
4QR5
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-[3-(cyclopentylsulfamoyl)-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)phenyl]cyclopropanecarboxamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
4QR3
 
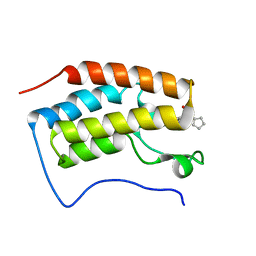 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | Bromodomain-containing protein 4, N-cyclopentyl-3-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
4QR4
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | 2-chloro-N-cyclopentyl-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
2WWP
 
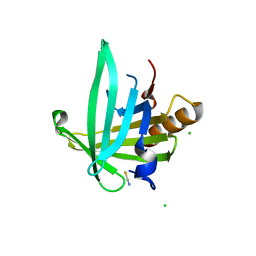 | | Crystal structure of the human lipocalin-type prostaglandin D synthase | | Descriptor: | CHLORIDE ION, PROSTAGLANDIN-H2 D-ISOMERASE, THIOCYANATE ION | | Authors: | Roos, A.K, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotyenova, T, Kotzch, A, Kraulis, P, Markova, N, Moche, M, Nielsen, T.K, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Dynamic Insights Into Substrate Binding and Catalysis of Human Lipocalin Prostaglandin D Synthase.
J.Lipid Res., 54, 2013
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7X2H
 
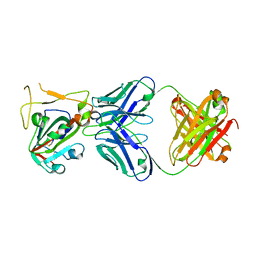 | |
7WP8
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK1628x in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7XD2
 
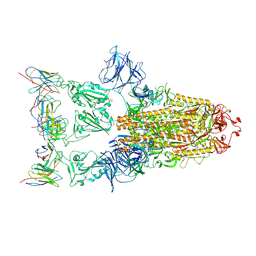 | |
5NPP
 
 | | 2.22A STRUCTURE OF THIOPHENE2 AND GSK945237 WITH S.AUREUS DNA GYRASE AND DNA | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, DIMETHYL SULFOXIDE, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Chan, P.F, Stavenger, R.A. | | Deposit date: | 2017-04-18 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Thiophene antibacterials that allosterically stabilize DNA-cleavage complexes with DNA gyrase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
