4OCK
 
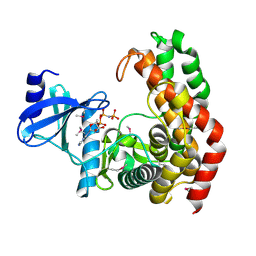 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc and AMPPNP | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCP
 
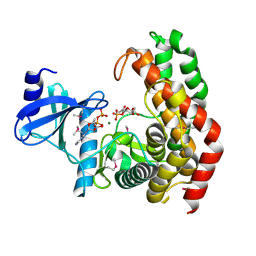 | | N-acetylhexosamine 1-phosphate kinase in complex with GlcNAc-1-phosphate and ADP | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OCV
 
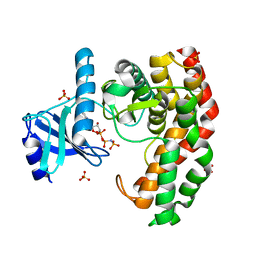 | | N-acetylhexosamine 1-phosphate kinase_ATCC15697 in complex with GlcNAc and AMPPNP | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, N-acetylhexosamine 1-phosphate kinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, T.L, Wang, K.C, Lyu, S.Y, Liu, Y.C, Chang, C.Y, Wu, C.J. | | Deposit date: | 2014-01-09 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Insights into the binding specificity and catalytic mechanism of N-acetylhexosamine 1-phosphate kinases through multiple reaction complexes.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6NIE
 
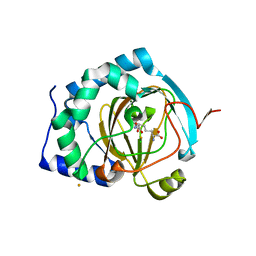 | | BesD with Fe(II), chloride, and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, BesD, lysine halogenase, ... | | Authors: | Neugebauer, M.E, Chang, M.C.Y. | | Deposit date: | 2018-12-27 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A family of radical halogenases for the engineering of amino-acid-based products.
Nat.Chem.Biol., 15, 2019
|
|
7TWA
 
 | | Crystal structure of apo BesC from Streptomyces cattleya | | Descriptor: | 1,3-BUTANEDIOL, 4-chloro-allylglycine synthase, ACETATE ION, ... | | Authors: | Neugebauer, M.E, McBride, M.J, Boal, A.K, Chang, M.C.Y. | | Deposit date: | 2022-02-07 | | Release date: | 2022-04-13 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate-Triggered mu-Peroxodiiron(III) Intermediate in the 4-Chloro-l-Lysine-Fragmenting Heme-Oxygenase-like Diiron Oxidase (HDO) BesC: Substrate Dissociation from, and C4 Targeting by, the Intermediate.
Biochemistry, 61, 2022
|
|
7U6J
 
 | | HalB with lysine and succinate | | Descriptor: | Halogenase B, LYSINE, SUCCINIC ACID | | Authors: | Sumida, K.H, Neugebauer, M.E, Kissman, E.N, Chang, M.C.Y. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biocatalytic control of site-selectivity and chain length-selectivity in radical amino acid halogenases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7U6H
 
 | | HalD with ornithine and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Swenson, C.V, Neugebauer, M.E, Kissman, E.N, Chang, M.C.Y. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biocatalytic control of site-selectivity and chain length-selectivity in radical amino acid halogenases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7U6I
 
 | | HalB with glycine and succinate | | Descriptor: | GLYCEROL, GLYCINE, Halogenase B, ... | | Authors: | Neugebauer, M.E, Kissman, E.N, Chang, M.C.Y. | | Deposit date: | 2022-03-04 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Biocatalytic control of site-selectivity and chain length-selectivity in radical amino acid halogenases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7JSD
 
 | | Hydroxylase homolog of BesD with Fe(II), alpha-ketoglutarate, and lysine | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, LYSINE, ... | | Authors: | Kissman, E.N, Neugebauer, M.E, Chang, M.C.Y. | | Deposit date: | 2020-08-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reaction pathway engineering converts a radical hydroxylase into a halogenase.
Nat.Chem.Biol., 18, 2022
|
|
6BJ9
 
 | |
6BJA
 
 | |
6BJB
 
 | |
4R82
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 in complex with NAD and FAD fragments | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-01 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
4HX6
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 | | Descriptor: | ACETATE ION, Oxidoreductase, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-09 | | Release date: | 2012-11-28 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
4OO2
 
 | | Streptomyces globisporus C-1027 FAD dependent (S)-3-chloro-β-tyrosine-S-SgcC2 C-5 hydroxylase SgcC apo form | | Descriptor: | CALCIUM ION, Chlorophenol-4-monooxygenase, GLYCEROL | | Authors: | Cao, H, Xu, W, Bingman, C.A, Lohman, J.R, Yennamalli, R, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-01-29 | | Release date: | 2014-02-12 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
2REW
 
 | | Crystal Structure of PPARalpha ligand binding domain with BMS-631707 | | Descriptor: | (2S,3S)-1-(4-METHOXYPHENYL)-3-(3-(2-(5-METHYL-2-PHENYLOXAZOL-4-YL)ETHOXY)BENZYL)-4-OXOAZETIDINE-2-CARBOXYLIC ACID, N,N-BIS(3-D-GLUCONAMIDOPROPYL)DEOXYCHOLAMIDE, Peroxisome proliferator-activated receptor alpha | | Authors: | Muckelbauer, J. | | Deposit date: | 2007-09-27 | | Release date: | 2007-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Azetidinone Acids as Conformationally-Constrained Dual (alpha/gamma) PPAR Activators
To be Published
|
|
7YPU
 
 | | OrfE-CoA-glycylthricin complex | | Descriptor: | Acetyltransferase, COENZYME A, [(2~{R},3~{R},4~{S},5~{R},6~{R})-6-[(~{E})-[(3~{a}~{S},7~{R},7~{a}~{S})-7-oxidanyl-4-oxidanylidene-3,3~{a},5,6,7,7~{a}-hexahydro-1~{H}-imidazo[4,5-c]pyridin-2-ylidene]amino]-5-(2-azanylethanoylamino)-2-(hydroxymethyl)-4-oxidanyl-oxan-3-yl] carbamate | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-08-04 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
7YPV
 
 | | Crystal structure of OrE-ST-F | | Descriptor: | Acetyltransferase, Streptothricin F | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-08-04 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
4Q38
 
 | | The crystal structure of acyltransferase in complex with decanoyl-CoA and teicoplanin | | Descriptor: | 2-amino-6-O-decanoyl-2-deoxy-beta-D-glucopyranose, COENZYME A, Putative uncharacterized protein tcp24, ... | | Authors: | Li, T.-L, Lyu, S.-Y, Liu, Y.-C, Chang, C.-Y, Huang, C.-J. | | Deposit date: | 2014-04-11 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
4Q36
 
 | | The crystal structure of acyltransferase in complex with octanoyl-CoA and teicoplanin | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, OCTANOYL-COENZYME A, Putative uncharacterized protein tcp24, ... | | Authors: | Li, T.-L, Lyu, S.-Y, Liu, Y.-C, Chang, C.-Y, Huang, C.-J. | | Deposit date: | 2014-04-11 | | Release date: | 2014-09-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Multiple complexes of long aliphatic N-acyltransferases lead to synthesis of 2,6-diacylated/2-acyl-substituted glycopeptide antibiotics, effectively killing vancomycin-resistant enterococcus
J.Am.Chem.Soc., 136, 2014
|
|
8GRI
 
 | | Orf1-E312A-glycine-glycylthricin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCINE, N-formimidoyl fortimicin A synthase, ... | | Authors: | Wang, Y.L, Li, T.L. | | Deposit date: | 2022-09-01 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.365 Å) | | Cite: | N-Formimidoylation/-iminoacetylation modification in aminoglycosides requires FAD-dependent and ligand-protein NOS bridge dual chemistry.
Nat Commun, 14, 2023
|
|
3K5U
 
 | | Identification, SAR Studies and X-ray Cocrystal Analysis of a Novel Furano-pyrimidine Aurora Kinase A Inhibitor | | Descriptor: | 2-[(5,6-DIPHENYLFURO[2,3-D]PYRIMIDIN-4-YL)AMINO]ETHANOL, Serine/threonine-protein kinase 6 | | Authors: | Wu, J.S, Leou, J.S, Coumar, M.S, Hsieh, H.P, Wu, S.Y. | | Deposit date: | 2009-10-08 | | Release date: | 2010-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification, SAR studies, and X-ray co-crystallographic analysis of a novel furanopyrimidine aurora kinase A inhibitor
Chemmedchem, 5, 2010
|
|
5IUG
 
 | | Crystal Structure of Anaplastic Lymphoma Kinase (ALK) in complex with 5a | | Descriptor: | ALK tyrosine kinase receptor, N-[3-(4-{[(5-tert-butyl-1,2-oxazol-3-yl)carbamoyl]amino}-3-methylphenyl)-1H-pyrazol-5-yl]-4-[(4-methylpiperazin-1-yl)methyl]benzamide | | Authors: | Tu, C.H, Wu, S.Y. | | Deposit date: | 2016-03-18 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Pyrazolylamine Derivatives Reveal the Conformational Switching between Type I and Type II Binding Modes of Anaplastic Lymphoma Kinase (ALK).
J.Med.Chem., 59, 2016
|
|
5IUH
 
 | | Crystal Structure of the Anaplastic Lymphoma Kinase (ALK) in complex with 5d | | Descriptor: | 4-[(4-methylpiperazin-1-yl)methyl]-N-{3-[3-methyl-4-({[5-(propan-2-yl)-1,2-oxazol-3-yl]carbamoyl}amino)phenyl]-1H-pyrazol-5-yl}benzamide, ALK tyrosine kinase receptor | | Authors: | Tu, C.H, Wu, S.Y. | | Deposit date: | 2016-03-18 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyrazolylamine Derivatives Reveal the Conformational Switching between Type I and Type II Binding Modes of Anaplastic Lymphoma Kinase (ALK).
J.Med.Chem., 59, 2016
|
|
5IUI
 
 | | Crystal Structure of Anaplastic Lyphoma Kinase (ALK) in complex with 4 | | Descriptor: | ALK tyrosine kinase receptor, N-[3-(4-amino-3-methylphenyl)-1H-pyrazol-5-yl]-4-[(4-methylpiperazin-1-yl)methyl]benzamide | | Authors: | Tu, C.H, Wu, S.Y. | | Deposit date: | 2016-03-18 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Pyrazolylamine Derivatives Reveal the Conformational Switching between Type I and Type II Binding Modes of Anaplastic Lymphoma Kinase (ALK).
J.Med.Chem., 59, 2016
|
|
