4E4K
 
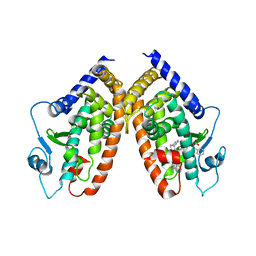 | | Crystal Structure of PPARgamma with the ligand JO21 | | Descriptor: | (2S)-3-phenyl-2-{[2'-(propan-2-yl)biphenyl-4-yl]oxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Loiodice, F, Fracchiolla, G, Laghezza, A, Carbonara, G, Piemontese, L, Lavecchia, A, Novellino, E. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New 2-(Aryloxy)-3-phenylpropanoic Acids as Peroxisome Proliferator-Activated Receptor alpha/gamma Dual Agonists Able To Upregulate Mitochondrial Carnitine Shuttle System Gene Expression.
J.Med.Chem., 56, 2013
|
|
4E4Q
 
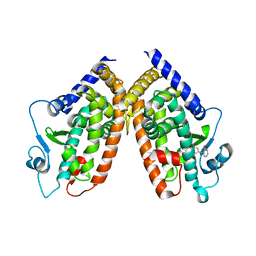 | | Crystal structure of PPARgamma with the ligand FS214 | | Descriptor: | (2R)-3-phenyl-2-{[2'-(propan-2-yl)biphenyl-4-yl]oxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Loiodice, F, Fracchiolla, G, Laghezza, A, Carbonara, G, Piemontese, L, Lavecchia, A, Novellino, E. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New 2-(Aryloxy)-3-phenylpropanoic Acids as Peroxisome Proliferator-Activated Receptor alpha/gamma Dual Agonists Able To Upregulate Mitochondrial Carnitine Shuttle System Gene Expression.
J.Med.Chem., 56, 2013
|
|
7DD7
 
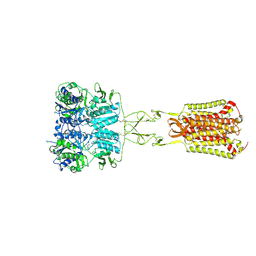 | | Structure of Calcium-Sensing Receptor in complex with Evocalcet | | Descriptor: | 2-[4-[(3S)-3-[[(1R)-1-naphthalen-1-ylethyl]amino]pyrrolidin-1-yl]phenyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wen, T.L, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-16 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7DD5
 
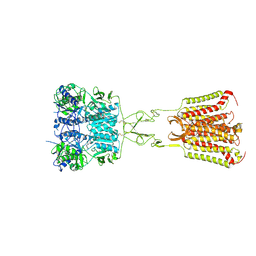 | | Structure of Calcium-Sensing Receptor in complex with NPS-2143 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, ... | | Authors: | Wen, T.L, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-16 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7DD6
 
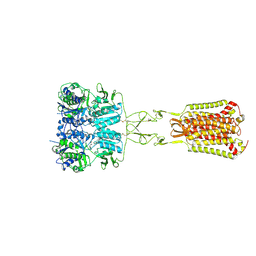 | | Structure of Ca2+/L-Trp-bonnd Calcium-Sensing Receptor in active state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wen, T.L, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-16 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7EBS
 
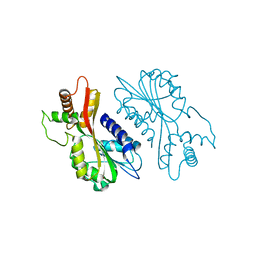 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT from silkworm | | Descriptor: | Juvenile hormone acid methyltransferase | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
7EBX
 
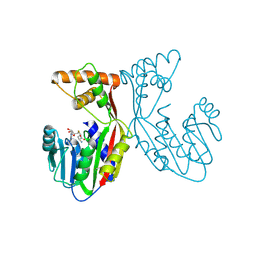 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT in complex with S-adenosyl-L-homocysteine. | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, juvenile hormone acid methyltransferase | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
7E4M
 
 | |
7EC0
 
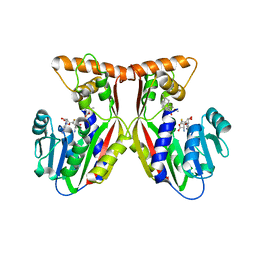 | | Crystal structure of juvenile hormone acid methyltransferase JHAMT in complex with S-Adenosyl homocysteine and methyl farnesoate | | Descriptor: | Juvenile hormone acid methyltransferase, Methyl farnesoate, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Guo, P.C, Zhang, Y.S, Zhang, L, Xu, H.Y, Xia, Q.Y. | | Deposit date: | 2021-03-11 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structural basis for juvenile hormone biosynthesis by the juvenile hormone acid methyltransferase.
J.Biol.Chem., 297, 2021
|
|
7E4O
 
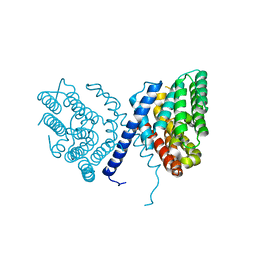 | |
7E4N
 
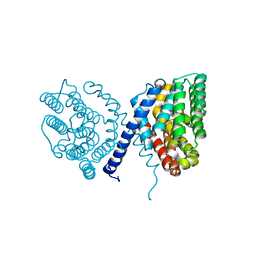 | | Crystal structure of Sat1646 | | Descriptor: | MAGNESIUM ION, Sat1646 | | Authors: | Yu, J.H, Xing, B.Y, Ma, M. | | Deposit date: | 2021-02-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Functional characterization and structural bases of two class I diterpene synthases in pimarane-type diterpene biosynthesis
Commun Chem, 4, 2021
|
|
7E53
 
 | |
7DUY
 
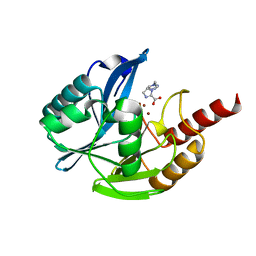 | | Crystal structure of VIM-2 MBL in complex with 1-(2-(1H-1,2,3-triazol-1-yl)ethyl)-1H-imidazole-2-carboxylic acid | | Descriptor: | 1-[2-(1,2,3-triazol-1-yl)ethyl]imidazole-2-carboxylic acid, Beta-lactamase class B VIM-2, ZINC ION | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2021-01-12 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure-guided optimization of 1H-imidazole-2-carboxylic acid derivatives affording potent VIM-Type metallo-beta-lactamase inhibitors.
Eur.J.Med.Chem., 228, 2022
|
|
7DV0
 
 | |
7DUX
 
 | |
7DU1
 
 | |
7DUE
 
 | |
7DUB
 
 | |
7DV1
 
 | |
7DUZ
 
 | |
7DYY
 
 | |
7DYZ
 
 | |
7DZ1
 
 | |
7DZ0
 
 | |
7DM2
 
 | | crystal structure of the M. tuberculosis phosphate ABC transport receptor PstS-1 in complex with Fab p4-170 | | Descriptor: | PHOSPHATE ION, Phosphate-binding protein PstS 1, heavy chain, ... | | Authors: | Ma, B, Freund, N, Xiang, Y. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human antibodies targeting a Mycobacterium transporter protein mediate protection against tuberculosis.
Nat Commun, 12, 2021
|
|
