5HXD
 
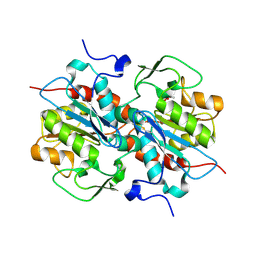 | | Crystal structure of murein-tripeptide amidase MpaA from Escherichia coli O157 | | Descriptor: | CACODYLATE ION, Protein MpaA, ZINC ION | | Authors: | Ma, Y, Bai, G, Zhang, X, Zhao, J, Yuan, Z, Kang, X, Li, Z, Mu, S, Liu, X. | | Deposit date: | 2016-01-30 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Murein-Tripeptide Amidase MpaA from Escherichia coli O157 at 2.6 angstrom Resolution
Protein Pept.Lett., 24, 2017
|
|
5ZU6
 
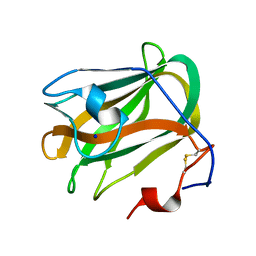 | | A CBM32 derived from alginate lyase B (AlyB-OU02) | | Descriptor: | CBM32 domain, SODIUM ION | | Authors: | Liu, W, Lyu, Q, Zhang, K. | | Deposit date: | 2018-05-07 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and biochemical characterization of a multidomain alginate lyase reveals a novel role of CBM32 in CAZymes
Biochim. Biophys. Acta, 1862, 2018
|
|
6EJG
 
 | |
6E7G
 
 | |
8JGD
 
 | | GDP-bound KRAS G12C in complex with YK-8S | | Descriptor: | (2~{S})-1-[4-[7-(8-ethynyl-7-fluoranyl-naphthalen-1-yl)-8-fluoranyl-2-[[(2~{R},8~{S})-2-fluoranyl-1,2,3,5,6,7-hexahydropyrrolizin-8-yl]methoxy]pyrido[4,3-d]pyrimidin-4-yl]piperazin-1-yl]-2-oxidanyl-propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z.M, Wang, R.L. | | Deposit date: | 2023-05-20 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.60037053 Å) | | Cite: | Simultaneous Covalent Modification of K-Ras(G12D) and K-Ras(G12C) with Tunable Oxirane Electrophiles.
J.Am.Chem.Soc., 145, 2023
|
|
8JHL
 
 | | GDP-bound KRAS G12D in complex with YK-8S | | Descriptor: | 1-[4-[7-(8-ethynyl-7-fluoranyl-naphthalen-1-yl)-8-fluoranyl-2-[[(2~{R},8~{S})-2-fluoranyl-1,2,3,5,6,7-hexahydropyrrolizin-8-yl]methoxy]pyrido[4,3-d]pyrimidin-4-yl]piperazin-1-yl]-3-oxidanyl-propan-1-one, GTPase KRas, N-terminally processed, ... | | Authors: | Zhang, Z.M, Wang, R.L. | | Deposit date: | 2023-05-23 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.10004044 Å) | | Cite: | Simultaneous Covalent Modification of K-Ras(G12D) and K-Ras(G12C) with Tunable Oxirane Electrophiles.
J.Am.Chem.Soc., 145, 2023
|
|
4Y8C
 
 | | Crystal structure of phosphodiesterase 9 in complex with (S)-C33 | | Descriptor: | 6-{[(1S)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Huang, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
6E24
 
 | | Ternary structure of c-Myc-TBP-TAF1 | | Descriptor: | Transcription initiation factor TFIID subunit 1,Myc proto-oncogene protein,TATA-box-binding protein | | Authors: | Wei, Y, Dong, A, Sunnerhagen, M, Penn, L, Tong, Y, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-07-10 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Multiple direct interactions of TBP with the MYC oncoprotein.
Nat.Struct.Mol.Biol., 26, 2019
|
|
1QXF
 
 | | SOLUTION STRUCTURE OF 30S RIBOSOMAL PROTEIN S27E FROM ARCHAEOGLOBUS FULGIDUS: GR2, A NESG TARGET PROTEIN | | Descriptor: | 30S RIBOSOMAL PROTEIN S27E | | Authors: | Herve Du Penhoat, C, Atreya, H.S, Shen, Y, Liu, G, Acton, T.B, Xiao, R, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of the 30S ribosomal protein S27e encoded in gene RS27_ARCFU of Archaeoglobus fulgidis reveals a novel protein fold
Protein Sci., 13, 2004
|
|
6EJM
 
 | |
6EK2
 
 | |
6EKG
 
 | |
6E7I
 
 | | Human ppGalNAcT2 I253A/L310A Mutant with EA2 and UDP | | Descriptor: | EA2, MANGANESE (II) ION, Polypeptide N-acetylgalactosaminyltransferase 2, ... | | Authors: | Bertozzi, C.R, Schumann, B, Agbay, A.J. | | Deposit date: | 2018-07-26 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bump-and-Hole Engineering Identifies Specific Substrates of Glycosyltransferases in Living Cells.
Mol.Cell, 78, 2020
|
|
7WK2
 
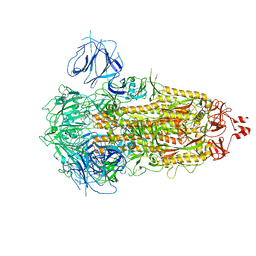 | | SARS-CoV-2 Omicron S-close | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WVO
 
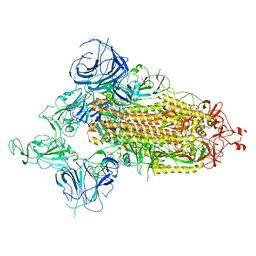 | | SARS-CoV-2 Omicron S-open-2 | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
7WVN
 
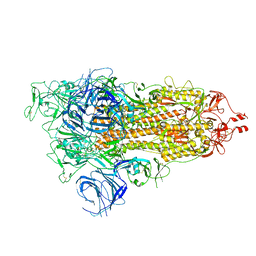 | | SARS-CoV-2 Omicron S-open | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-03-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of receptor binding and antibody neutralization of Omicron.
Nature, 604, 2022
|
|
4Y87
 
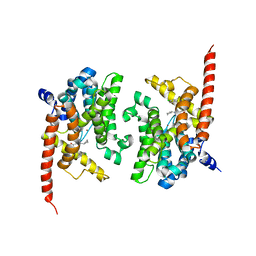 | | Crystal structure of phosphodiesterase 9 in complex with (R)-C33 (6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one) | | Descriptor: | 6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Huang, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
6EKH
 
 | |
4WYH
 
 | |
6E7H
 
 | |
4Y86
 
 | | Crystal structure of PDE9 in complex with racemic inhibitor C33 | | Descriptor: | 6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, 6-{[(1S)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, ... | | Authors: | Huang, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-09-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
6E16
 
 | | Ternary structure of c-Myc-TBP-TAF1 | | Descriptor: | Transcription initiation factor TFIID subunit 1,Myc proto-oncogene protein,TATA-box-binding protein | | Authors: | Wei, Y, Dong, A, Sunnerhagen, M, Penn, L, Tong, Y, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-07-09 | | Release date: | 2019-10-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multiple direct interactions of TBP with the MYC oncoprotein.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6BYO
 
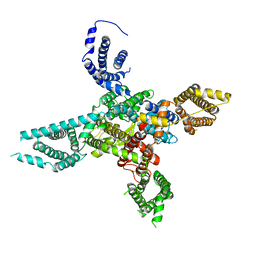 | |
6DDJ
 
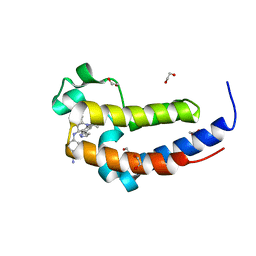 | | Crystal Structure of the human BRD2 BD2 bromodimain in complex with a Tetrahydroquinoline analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2S,4R)-1-acetyl-2-methyl-6-(1H-pyrazol-3-yl)-1,2,3,4-tetrahydroquinolin-4-yl]amino}benzonitrile, Bromodomain-containing protein 2 | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2018-05-10 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Bromodomain-Selective BET Inhibitors Are Potent Antitumor Agents against MYC-Driven Pediatric Cancer.
Cancer Res., 80, 2020
|
|
8XDA
 
 | | Cryo-EM structure of urea bound human urea transporter A2. | | Descriptor: | UREA, Urea transporter 2 | | Authors: | Huang, S, Liu, L, Sun, J, Zhizheng, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the mechanisms of urea permeation and distinct inhibition modes of urea transporters.
Nat Commun, 15, 2024
|
|
