5UN0
 
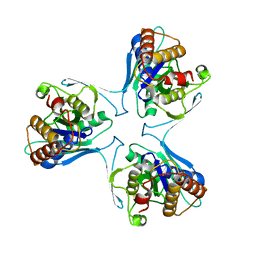 | | Crystal Structure of Mycobacterium Tuberculosis Proteasome-assembly chaperone homologue Rv2125 | | Descriptor: | proteasome assembly chaperone 2 (PAC2) homologue Rv2125 | | Authors: | Bai, L, Jastrab, J.B, Hu, K, Yu, H, Darwin, K.H, Li, H. | | Deposit date: | 2017-01-30 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of Mycobacterium tuberculosis Homologues of the Eukaryotic Proteasome Assembly Chaperone 2 (PAC2).
J. Bacteriol., 199, 2017
|
|
1K2Z
 
 | |
5UQC
 
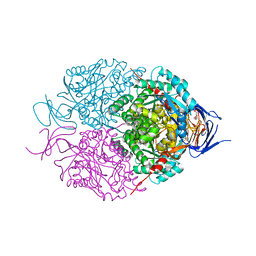 | | Crystal structure of mouse CRMP2 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, Dihydropyrimidinase-related protein 2 | | Authors: | Khanna, M, Khanna, R, Perez-Miller, S, Francois-Moutal, L. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A single structurally conserved SUMOylation site in CRMP2 controls NaV1.7 function.
Channels (Austin), 11, 2017
|
|
5URK
 
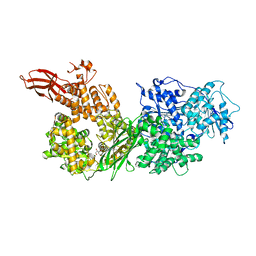 | | Crystal structure of human BRR2 in complex with T-3935799 | | Descriptor: | 6-benzyl-3-[3-(benzyloxy)phenyl]-4,6-dihydropyrido[4,3-d]pyrimidine-2,7(1H,3H)-dione, GLYCEROL, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Qin, L, Tjhen, R, Klein, M.G. | | Deposit date: | 2017-02-10 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Discovery of Allosteric Inhibitors Targeting the Spliceosomal RNA Helicase Brr2.
J. Med. Chem., 60, 2017
|
|
1JPY
 
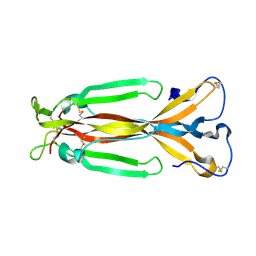 | | Crystal structure of IL-17F | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hymowitz, S.G, Filvaroff, E.H, Yin, J, Lee, J, Cai, L, Risser, P, Maruoka, M, Mao, W, Foster, J, Kelley, R, Pan, G, Gurney, A.L, de Vos, A.M, Starovasnik, M.A. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | IL-17s adopt a cystine knot fold: structure and activity of a novel cytokine, IL-17F, and implications for receptor binding.
EMBO J., 20, 2001
|
|
1JG7
 
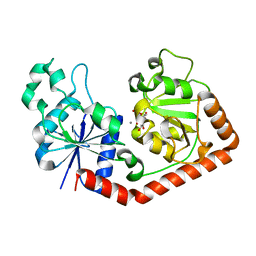 | | T4 phage BGT in complex with UDP and Mn2+ | | Descriptor: | DNA BETA-GLUCOSYLTRANSFERASE, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Morera, S, Lariviere, L, Kurzeck, J, Aschke-Sonnenborn, U, Freemont, P.S, Janin, J, Ruger, W. | | Deposit date: | 2001-06-23 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding.
J.Mol.Biol., 311, 2001
|
|
1JSP
 
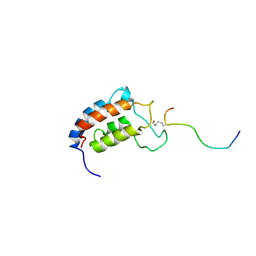 | | NMR Structure of CBP Bromodomain in complex with p53 peptide | | Descriptor: | CREB-BINDING PROTEIN, tumor protein p53 | | Authors: | He, Y, Mujtaba, S, Zeng, L, Yan, S, Zhou, M.-M. | | Deposit date: | 2001-08-17 | | Release date: | 2002-08-17 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation.
Mol.Cell, 13, 2004
|
|
7CRD
 
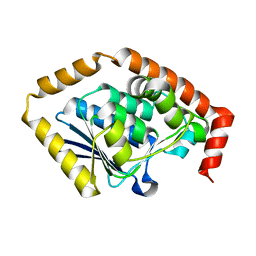 | | Structure of Pseudomonas aeruginosa OdaA | | Descriptor: | Probable enoyl-CoA hydratase/isomerase | | Authors: | Zhao, N, Zhao, C, Liu, L, Li, T, Li, C, He, L, Zhu, Y, Song, Y, Bao, R. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structural and molecular dynamic studies of Pseudomonas aeruginosa OdaA reveal the regulation role of a C-terminal hinge element.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
1JJI
 
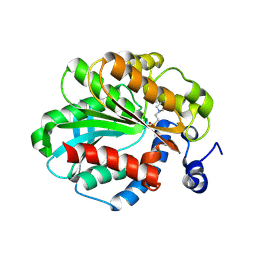 | | The Crystal Structure of a Hyper-thermophilic Carboxylesterase from the Archaeon Archaeoglobus fulgidus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Carboxylesterase | | Authors: | De Simone, G, Menchise, V, Manco, G, Mandrich, L, Sorrentino, N, Lang, D, Rossi, M, Pedone, C. | | Deposit date: | 2001-07-06 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a hyper-thermophilic carboxylesterase from the archaeon Archaeoglobus fulgidus.
J.Mol.Biol., 314, 2001
|
|
1NM4
 
 | | Solution structure of Cu(I)-CopC from Pseudomonas syringae | | Descriptor: | Copper resistance protein C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Mangani, S, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-01-09 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A redox switch in CopC: An intriguing copper trafficking protein that binds copper(I) and copper(II)
at different sites
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5UVK
 
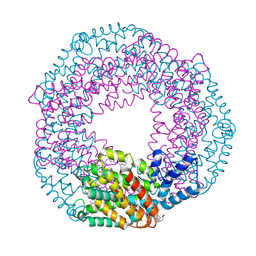 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
Iucrj, 4, 2017
|
|
1K06
 
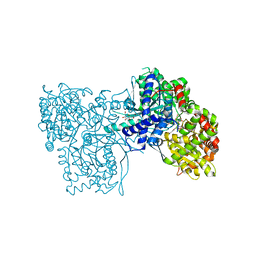 | | Crystallographic Binding Study of 100 mM N-benzoyl-N'-beta-D-glucopyranosyl urea to glycogen phosphorylase b | | Descriptor: | Glycogen Phosphorylase, N-[(phenylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Oikonomakos, N.G, Kosmopoulou, M, Zographos, S.E, Leonidas, D.D, Chrysina, E.D, Somsak, L, Nagy, V, Praly, J.P, Docsa, T, Toth, B, Gergely, P. | | Deposit date: | 2001-09-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding of N-acetyl-N '-beta-D-glucopyranosyl urea and N-benzoyl-N '-beta-D-glucopyranosyl urea to glycogen phosphorylase b: kinetic and crystallographic studies.
Eur.J.Biochem., 269, 2002
|
|
1NPU
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MURINE PD-1 | | Descriptor: | Programmed cell death protein 1 | | Authors: | Zhang, X, Schwartz, J.-C.D, Guo, X, Cao, E, Chen, L, Zhang, Z.-Y, Nathenson, S.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-20 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the costimulatory receptor programmed death-1.
Immunity, 20, 2004
|
|
5V23
 
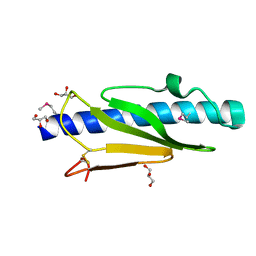 | |
1JUF
 
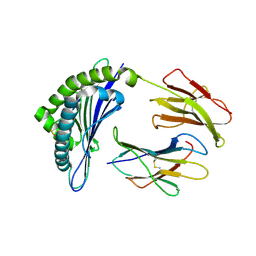 | | Structure of Minor Histocompatibility Antigen peptide, H13b, complexed to H2-Db | | Descriptor: | Beta-2-microglobulin, H13b peptide, H2-Db major histocompatibility antigen | | Authors: | Ostrov, D.A, Roden, M.M, Shi, W, Palmieri, E, Christianson, G.J, Mendoza, L, Villaflor, G, Tilley, D, Shastri, N, Grey, H, Almo, S.C, Roopenian, D.C, Nathenson, S.G. | | Deposit date: | 2001-08-24 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How H13 histocompatibility peptides differing by a single methyl group and lacking conventional MHC binding anchor motifs determine self-nonself discrimination.
J.Immunol., 168, 2002
|
|
7RF6
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the second illumination at 2.01 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
7RMT
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-70 | | Descriptor: | 2-chloro-4-[4-(2,6-dioxo-1,2,5,6-tetrahydropyrimidine-4-carbonyl)piperazin-1-yl]benzaldehyde, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-28 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RNH
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-45 | | Descriptor: | 3C-like proteinase, 6-[4-(4-chlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RF4
 
 | | RT XFEL structure of Photosystem II 50 microseconds after the second illumination at 2.27 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
7RF2
 
 | | RT XFEL structure of dark-stable state of Photosystem II (0F, S1 rich) at 2.08 Angstrom | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Hussein, R, Ibrahim, M, Bhowmick, A, Simon, P.S, Chatterjee, R, Lassalle, L, Doyle, M.D, Bogacz, I, Kim, I.-S, Cheah, M.H, Gul, S, de Lichtenberg, C, Chernev, P, Pham, C.C, Young, I.D, Carbajo, S, Fuller, F.D, Alonso-Mori, R, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bolotovski, R, Mendez, D, Holton, J.M, Moriarty, N.W, Adams, P.D, Bergmann, U, Sauter, N.K, Dobbek, H, Messinger, J, Zouni, A, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural dynamics in the water and proton channels of photosystem II during the S 2 to S 3 transition.
Nat Commun, 12, 2021
|
|
7RLS
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-68 | | Descriptor: | 3C-like proteinase, 6-[4-(3,4,5-trichlorophenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
5UYO
 
 | | Solution NMR structure of the de novo mini protein HEEH_rd4_0097 | | Descriptor: | HEEH_rd4_0097 | | Authors: | Lemak, A, Rocklin, G.J, Houliston, S, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
5UZS
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P200 | | Descriptor: | 1,2-ETHANEDIOL, 3-(2-{[(4-chlorophenyl)carbamoyl]amino}propan-2-yl)-N-hydroxybenzene-1-carboximidamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-27 | | Release date: | 2017-03-22 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.367 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P200
To Be Published
|
|
7RNK
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with HL-3-71 | | Descriptor: | 3C-like proteinase, 6-{4-[3-chloro-4-(hydroxymethyl)phenyl]piperazine-1-carbonyl}pyrimidine-2,4(3H,5H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-29 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
7RM2
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (Mpro) in complex with Mcule-CSR-494190-S1 | | Descriptor: | 3C-like proteinase, 6-[4-(3,5-dichloro-4-methylphenyl)piperazine-1-carbonyl]pyrimidine-2,4(1H,3H)-dione | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2021-07-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease.
J.Med.Chem., 64, 2021
|
|
