6J5B
 
 | | Structural basis for the target DNA recognition and binding by the MYB domain of phosphate starvation response regulator 1 | | Descriptor: | DNA (5'-D(*GP*GP*TP*AP*CP*AP*GP*TP*AP*TP*AP*TP*AP*CP*CP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*GP*GP*TP*AP*TP*AP*TP*AP*CP*TP*GP*TP*AP*CP*C)-3'), Protein PHOSPHATE STARVATION RESPONSE 1 | | Authors: | Jiang, M.Q, Sun, L.F, Isupov, M.N, Wu, Y.K. | | Deposit date: | 2019-01-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the Target DNA recognition and binding by the MYB domain of phosphate starvation response 1.
Febs J., 286, 2019
|
|
7VRL
 
 | | Solution structure of Rbfox RRM bound to a non-cognate RNA | | Descriptor: | RNA (5'-R(*UP*GP*CP*AP*UP*AP*U)-3'), RNA binding protein fox-1 homolog 1 | | Authors: | Yang, F, Varani, G. | | Deposit date: | 2021-10-23 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Two distinct binding modes provide the RNA-binding protein RbFox with extraordinary sequence specificity.
Nat Commun, 14, 2023
|
|
7XBW
 
 | | Cryo-EM structure of the human chemokine receptor CX3CR1 in complex with Gi1 | | Descriptor: | CHOLESTEROL, CX3C chemokine receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Lu, M, Zhao, W, Han, S, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Activation of the human chemokine receptor CX3CR1 regulated by cholesterol.
Sci Adv, 8, 2022
|
|
7XBX
 
 | | Cryo-EM structure of the human chemokine receptor CX3CR1 in complex with CX3CL1 and Gi1 | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lu, M, Zhao, W, Han, S, Zhu, Y, Wu, B, Zhao, Q. | | Deposit date: | 2022-03-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Activation of the human chemokine receptor CX3CR1 regulated by cholesterol.
Sci Adv, 8, 2022
|
|
4KS9
 
 | | Crystal Structure of Malonyl-CoA decarboxylase (Rmet_2797) from Cupriavidus metallidurans, Northeast Structural Genomics Consortium Target CrR76 | | Descriptor: | MAGNESIUM ION, Malonyl-CoA decarboxylase | | Authors: | Forouhar, F, Tran, T.H, Lew, S, Seetharaman, J, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-17 | | Release date: | 2013-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of malonyl-coenzyme a decarboxylase provide insights into its catalytic mechanism and disease-causing mutations.
Structure, 21, 2013
|
|
3SOM
 
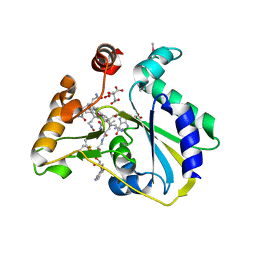 | | crystal structure of human MMACHC | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, 5'-DEOXYADENOSINE, ... | | Authors: | Krojer, T, Froese, D.S, von Delft, F, Muniz, J.R, Gileadi, C, Vollmar, M, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Gravel, R.A, Yue, W.W, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-30 | | Release date: | 2011-07-27 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of MMACHC reveals an arginine-rich pocket and a domain-swapped dimer for its B12 processing function.
Biochemistry, 51, 2012
|
|
4KSA
 
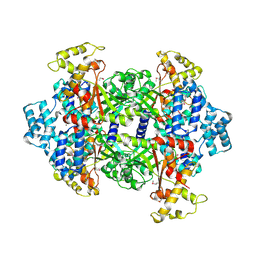 | | Crystal Structure of Malonyl-CoA decarboxylase from Rhodopseudomonas palustris, Northeast Structural Genomics Consortium Target RpR127 | | Descriptor: | MAGNESIUM ION, Malonyl-CoA decarboxylase | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Sahdev, S, Xiao, R, Patel, D.J, Ciccosanti, C, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-17 | | Release date: | 2013-06-19 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of malonyl-coenzyme a decarboxylase provide insights into its catalytic mechanism and disease-causing mutations.
Structure, 21, 2013
|
|
6KKU
 
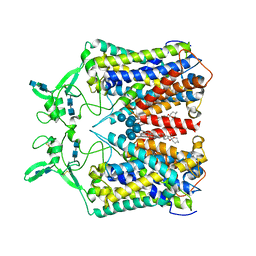 | | human KCC1 structure determined in NaCl and GDN | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Liu, S, Chang, S, Ye, S, Bai, X, Guo, J. | | Deposit date: | 2019-07-27 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of the human cation-chloride cotransporter KCC1.
Science, 366, 2019
|
|
7FHN
 
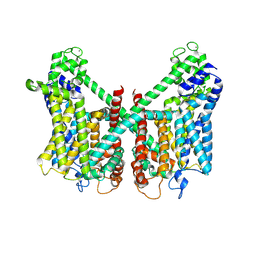 | | Structure of AtTPC1 D240A/D454A/E528A mutant with 1 mM Ca2+ | | Descriptor: | CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FHO
 
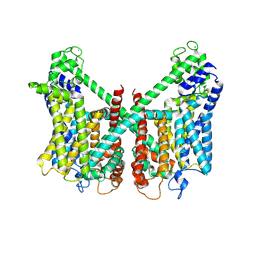 | | Structure of AtTPC1 D240A/D454A/E528A mutant with 50 mM Ca2+ | | Descriptor: | CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7FHL
 
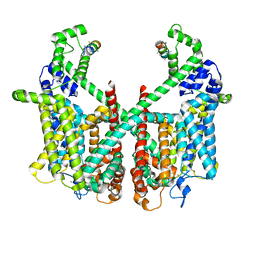 | | Structure of AtTPC1 with 50 mM Ca2+ | | Descriptor: | AtTPC1-Cter, CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ES2
 
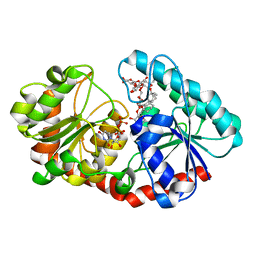 | | a mutant of glycosyktransferase in complex with UDP and Reb D | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, rebaudioside D | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7FHK
 
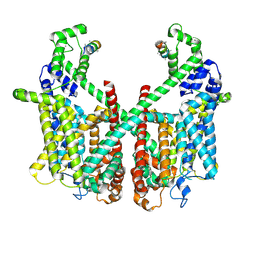 | | Structure of AtTPC1 with 1 mM Ca2+ | | Descriptor: | AtTPC1-Cter, CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ERY
 
 | | apo form of the glycosyltransferase | | Descriptor: | Glycosyltransferase | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ERX
 
 | | Glycosyltransferase in complex with UDP and STB | | Descriptor: | GLYCEROL, Glycosyltransferase, Steviolbioside, ... | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ES1
 
 | | glycosyltransferase in complex with UDP and ST | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, steviol-19-o-glucoside | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ES0
 
 | | a rice glycosyltransferase in complex with UDP and REX | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, ... | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
6AUD
 
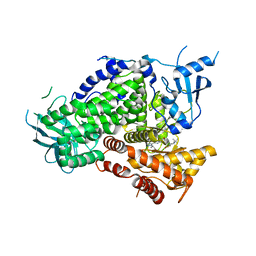 | | PI3K-gamma K802T in complex with Cpd 8 10-((1-(tert-butyl)piperidin-4-yl)sulfinyl)-2-(1-isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepine | | Descriptor: | 10-[(S)-(1-tert-butylpiperidin-4-yl)sulfinyl]-2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Ultsch, M. | | Deposit date: | 2017-08-31 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Design of Selective Benzoxazepin PI3K delta Inhibitors Through Control of Dihedral Angles.
ACS Med Chem Lett, 8, 2017
|
|
4KSF
 
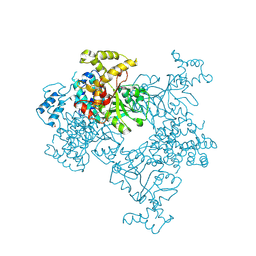 | | Crystal Structure of Malonyl-CoA decarboxylase from Agrobacterium vitis, Northeast Structural Genomics Consortium Target RiR35 | | Descriptor: | CHLORIDE ION, Malonyl-CoA decarboxylase, NICKEL (II) ION | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-17 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of malonyl-coenzyme a decarboxylase provide insights into its catalytic mechanism and disease-causing mutations.
Structure, 21, 2013
|
|
2IQF
 
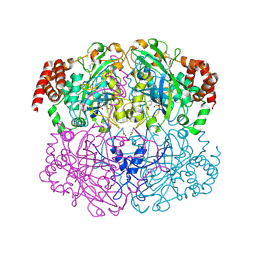 | | Crystal structure of Helicobacter pylori catalase compound I | | Descriptor: | ACETATE ION, Catalase, OXYGEN ATOM, ... | | Authors: | Loewen, P.C, Carpena, X, Fita, I. | | Deposit date: | 2006-10-13 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structures and electronic configuration of compound I intermediates of Helicobacter pylori and Penicillium vitale catalases determined by X-ray crystallography and QM/MM density functional theory calculations.
J.Am.Chem.Soc., 129, 2007
|
|
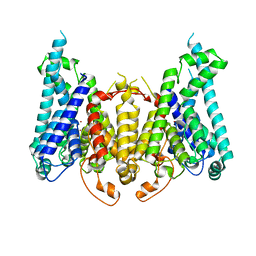
8W9T
 
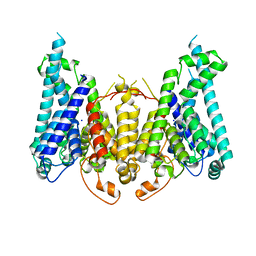 | |
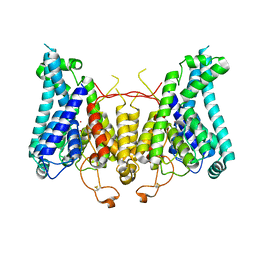
8W9V
 
 | |
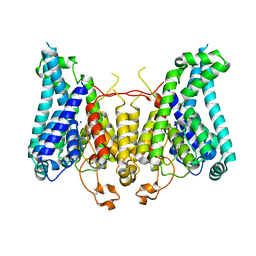
8W9O
 
 | |
8W9N
 
 | |
8V9Q
 
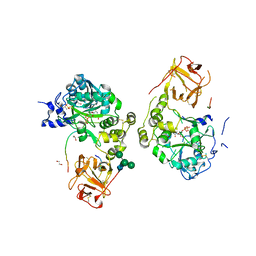 | | Crystal structure of mGalNAc-T1 in complex with the mucin glycopeptide Muc5AC-13, Mn2+, and UDP. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Samara, N.L, Collette, A.M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | An unusual dual sugar-binding lectin domain controls the substrate specificity of a mucin-type O-glycosyltransferase.
Sci Adv, 10, 2024
|
|
