8FO4
 
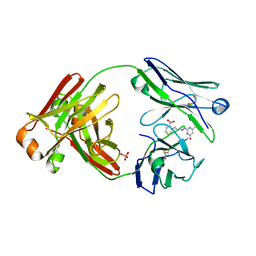 | |
1JGV
 
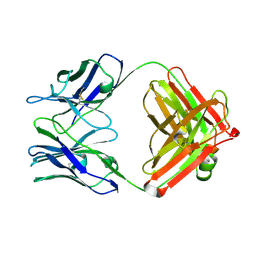 | | STRUCTURAL BASIS FOR DISFAVORED ELIMINATION REACTION IN CATALYTIC ANTIBODY 1D4 | | Descriptor: | Antibody Heavy Chain, Antibody Light Chain | | Authors: | Larsen, N.A, Heine, A, Crane, L, Cravatt, B.F, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2001-06-26 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for a disfavored elimination reaction in catalytic antibody 1D4.
J.Mol.Biol., 314, 2001
|
|
8FO5
 
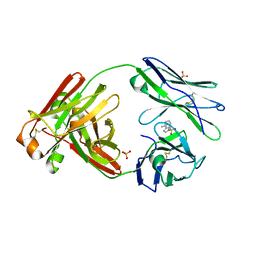 | |
1JKX
 
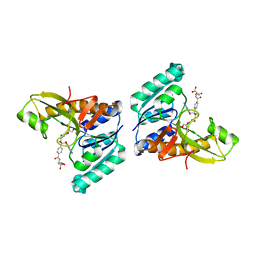 | | Unexpected formation of an epoxide-derived multisubstrate adduct inhibitor on the active site of GAR transformylase | | Descriptor: | N-[5'-O-PHOSPHONO-RIBOFURANOSYL]-2-[2-HYDROXY-2-[4-[GLUTAMIC ACID]-N-CARBONYLPHENYL]-3-[2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL]-PROPANYLAMINO]-ACETAMIDE, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Greasley, S.E, Marsilje, T.H, Cai, H, Baker, S, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2001-07-13 | | Release date: | 2001-11-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected formation of an epoxide-derived multisubstrate adduct inhibitor on the active site of GAR transformylase.
Biochemistry, 40, 2001
|
|
1FZO
 
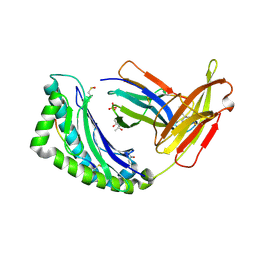 | | MHC CLASS I NATURAL MUTANT H-2KBM8 HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND SENDAI VIRUS NUCLEOPROTEIN | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rudolph, M.G, Speir, J.A, Brunmark, A, Mattsson, N, Jackson, M.R, Peterson, P.A, Teyton, L, Wilson, I.A. | | Deposit date: | 2000-10-03 | | Release date: | 2001-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of K(bm1) and K(bm8) reveal that subtle changes in the peptide environment impact thermostability and alloreactivity.
Immunity, 14, 2001
|
|
1JU3
 
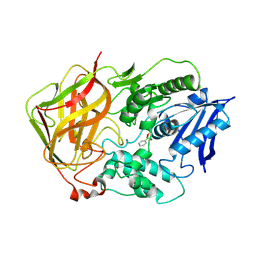 | | BACTERIAL COCAINE ESTERASE COMPLEX WITH TRANSITION STATE ANALOG | | Descriptor: | PHENYL BORONIC ACID, cocaine esterase | | Authors: | Larsen, N.A, Turner, J.M, Stevens, J, Rosser, S.J, Basran, A, Lerner, R.A, Bruce, N.C, Wilson, I.A. | | Deposit date: | 2001-08-23 | | Release date: | 2001-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a bacterial cocaine esterase.
Nat.Struct.Biol., 9, 2002
|
|
1KBG
 
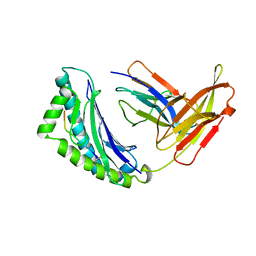 | | MHC Class I H-2KB Presented Glycopeptide RGY8-6H-GAL2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (BETA-2-MICROGLOBULIN), ... | | Authors: | Speir, J.A, Abdel-Motal, U.M, Jondal, M, Wilson, I.A. | | Deposit date: | 1998-08-28 | | Release date: | 1999-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an MHC class I presented glycopeptide that generates carbohydrate-specific CTL.
Immunity, 10, 1999
|
|
1JCJ
 
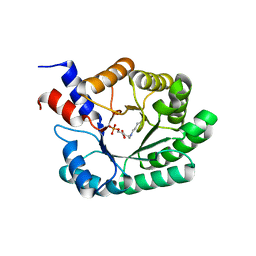 | | OBSERVATION OF COVALENT INTERMEDIATES IN AN ENZYME MECHANISM AT ATOMIC RESOLUTION | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, DEOXYRIBOSE-PHOSPHATE ALDOLASE | | Authors: | Heine, A, DeSantis, G, Luz, J.G, Mitchell, M, Wong, C.-H, Wilson, I.A. | | Deposit date: | 2001-06-09 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Observation of covalent intermediates in an enzyme mechanism at atomic resolution.
Science, 294, 2001
|
|
1J9C
 
 | | Crystal Structure of tissue factor-factor VIIa complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-phenylalaninamide, ... | | Authors: | Huang, M, Ruf, W, Edgington, T.S, Wilson, I.A. | | Deposit date: | 2001-05-24 | | Release date: | 2004-07-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand Induced Conformational Transitions of Tissue Factor. Crystal Structure of the Tissue Factor:Factor VIIa Complex.
To be Published
|
|
1JGU
 
 | | STRUCTURAL BASIS FOR DISFAVORED ELIMINATION REACTION IN CATALYTIC ANTIBODY 1D4 | | Descriptor: | (2-AMINO-3-PHENYL-BICYCLO[2.2.1]HEPT-2-YL)-PHENYL-METHANONE, Antibody Heavy Chain, Antibody Light Chain, ... | | Authors: | Larsen, N.A, Heine, A, Crane, L, Cravatt, B.F, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2001-06-26 | | Release date: | 2001-12-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for a disfavored elimination reaction in catalytic antibody 1D4.
J.Mol.Biol., 314, 2001
|
|
1RC0
 
 | | Human GAR Tfase complex structure with polyglutamated 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-4-(2,4-DIAMINO-6-OXO-1,6-DIHYDRO-PYRIMIDIN-5-YL)-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXY-ETHYL)-BUT-2-YL-BENZOYL}-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GLUTAMIC ACID, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
1RBQ
 
 | | Human GAR Tfase complex structure with 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHATE ION, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Human GAR Tfase complex structure with polyglutamated
10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid
To be Published
|
|
1PL0
 
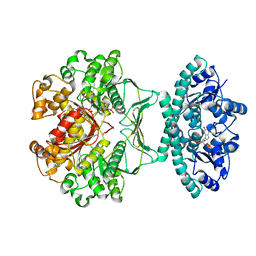 | | Crystal structure of human ATIC in complex with folate-based inhibitor, BW2315U89UC | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, N-(4-{[(2-AMINO-4-OXO-3,4-DIHYDROQUINAZOLIN-6-YL)AMINO]SULFONYL}BENZOYL)GLUTAMIC ACID, ... | | Authors: | Cheong, C.G, Greasley, S.E, Horton, P.A, Beardsley, G.P, Wilson, I.A. | | Deposit date: | 2003-06-06 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Human Bifunctional Enzyme Aminoimidazole-4-carboxamide Ribonucleotide Transformylase/IMP Cyclohydrolase in Complex with Potent Sulfonyl-containing Antifolates.
J.Biol.Chem., 279, 2004
|
|
1RC1
 
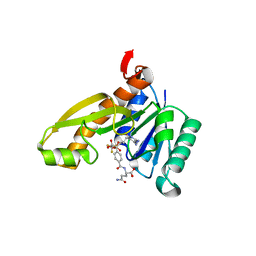 | | Human GAR Tfase complex structure with polyglutamated 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-4-(2,4-DIAMINO-6-OXO-1,6-DIHYDRO-PYRIMIDIN-5-YL)-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXY-ETHYL)-BUT-2-YL-BENZOYL}-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GLUTAMIC ACID, PHOSPHATE ION, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
1SQ2
 
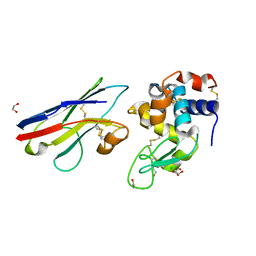 | | Crystal Structure Analysis of the Nurse Shark New Antigen Receptor (NAR) Variable Domain in Complex With Lysozyme | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Stanfield, R.L, Dooley, H, Flajnik, M.F, Wilson, I.A. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a shark single-domain antibody V region in complex with lysozyme.
Science, 305, 2004
|
|
1QWH
 
 | |
1RBZ
 
 | | Human GAR Tfase complex structure with polyglutamated 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-4-(2,4-DIAMINO-6-OXO-1,6-DIHYDRO-PYRIMIDIN-5-YL)-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXY-ETHYL)-BUT-2-YL-BENZOYL}-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GLUTAMIC ACID, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
1RD8
 
 | | Crystal Structure of the 1918 Human H1 Hemagglutinin Precursor (HA0) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stevens, J, Corper, A.L, Basler, C.F, Taubenberger, J.K, Palese, P, Wilson, I.A. | | Deposit date: | 2003-11-05 | | Release date: | 2004-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the Uncleaved Human H1 Hemagglutinin from the Extinct 1918 Influenza Virus.
Science, 303, 2004
|
|
1SJ5
 
 | | Crystal structure of a duf151 family protein (tm0160) from thermotoga maritima at 2.8 A resolution | | Descriptor: | conserved hypothetical protein TM0160 | | Authors: | Spraggon, G, Panatazatos, D, Klock, H.E, Wilson, I.A, Woods Jr, V.L, Lesley, S.A, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2004-03-02 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | On the use of DXMS to produce more crystallizable proteins: structures of the T. maritima proteins TM0160 and TM1171.
Protein Sci., 13, 2004
|
|
1SMO
 
 | | Crystal Structure of Human Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.47 . | | Descriptor: | L(+)-TARTARIC ACID, triggering receptor expressed on myeloid cells 1 | | Authors: | Kelker, M.S, Foss, T.R, Peti, W, Teyton, L, Kelly, J.W, Wilson, I.A. | | Deposit date: | 2004-03-09 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of Human Triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) at 1.47A.
J.Mol.Biol., 342, 2004
|
|
1R3H
 
 | | Crystal Structure of T10 | | Descriptor: | Beta-2-microglobulin, MHC H2-TL-T10-129 | | Authors: | Rudolph, M.G, Wilson, I.A. | | Deposit date: | 2003-10-02 | | Release date: | 2004-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Combined pseudo-merohedral twinning, non-crystallographic symmetry and pseudo-translation in a monoclinic crystal form of the gammadelta T-cell ligand T10.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1RBM
 
 | | Human GAR Tfase complex structure with polyglutamated 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-4-(2,4-DIAMINO-6-OXO-1,6-DIHYDRO-PYRIMIDIN-5-YL)-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXY-ETHYL)-BUT-2-YL-BENZOYL}-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GAMMA-GLUTAMYL-GLUTAMIC ACID, PHOSPHATE ION, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
1RBY
 
 | | Human GAR Tfase complex structure with 10-(trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic acid and substrate beta-GAR | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHORIBOSYLGLYCINAMIDE FORMYLTRANSFERASE | | Authors: | Zhang, Y, Desharnais, J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-11-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Human GAR Tfase complex structure
To be Published
|
|
1Q1J
 
 | | Crystal Structure Analysis of anti-HIV-1 Fab 447-52D in complex with V3 peptide | | Descriptor: | Fab 447-52D, heavy chain, light chain, ... | | Authors: | Stanfield, R.L, Gorny, M.K, Williams, C, Zolla-Pazner, S, Wilson, I.A. | | Deposit date: | 2003-07-21 | | Release date: | 2004-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D.
Structure, 12, 2004
|
|
1PKX
 
 | | Crystal Structure of human ATIC in complex with XMP | | Descriptor: | Bifunctional purine biosynthesis protein PURH, POTASSIUM ION, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Wolan, D.W, Cheong, C.G, Greasley, S.E, Wilson, I.A. | | Deposit date: | 2003-06-06 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Human and Avian IMP Cyclohydrolase Mechanism via Crystal Structures with the Bound XMP Inhibitor.
Biochemistry, 43, 2004
|
|
