3UV1
 
 | | Crystal structure a major allergen from dust mite | | Descriptor: | Der f 7 allergen | | Authors: | Tan, K.W, Kumar, T, Chew, F.T, Mok, Y.K. | | Deposit date: | 2011-11-29 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Der f 7, a Dust Mite Allergen from Dermatophagoides farinae.
Plos One, 7, 2012
|
|
3X12
 
 | | Crystal structure of HLA-B*57:01.I80N | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-57 alpha chain, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2014-10-24 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The interaction of KIR3DL1*001 with HLA class I molecules is dependent upon molecular microarchitecture within the Bw4 epitope
J.Immunol., 194, 2015
|
|
5B38
 
 | | KIR3DL1*005 in complex with HLA-B*57:01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2016-02-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition
J.Exp.Med., 213, 2016
|
|
3WYR
 
 | |
3X14
 
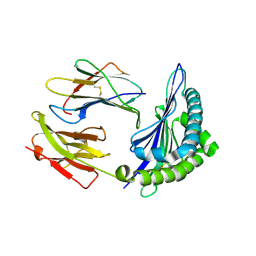 | | Crystal structure of HLA-B*0801.N80I.R82L.G83R | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-8 alpha chain, ... | | Authors: | Vivian, J.P, Rossjohn, J. | | Deposit date: | 2014-10-25 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The interaction of KIR3DL1*001 with HLA class I molecules is dependent upon molecular microarchitecture within the Bw4 epitope
J.Immunol., 194, 2015
|
|
1BF9
 
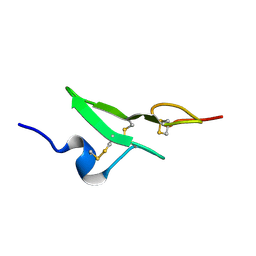 | | N-TERMINAL EGF-LIKE DOMAIN FROM HUMAN FACTOR VII, NMR, 23 STRUCTURES | | Descriptor: | FACTOR VII | | Authors: | Muranyi, A, Finn, B.E, Gippert, G.P, Forsen, S, Stenflo, J, Drakenberg, T. | | Deposit date: | 1998-05-28 | | Release date: | 1999-02-16 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal EGF-like domain from human factor VII.
Biochemistry, 37, 1998
|
|
6TLB
 
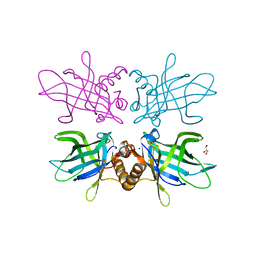 | | Plasmodium falciparum lipocalin (PF3D7_0925900) | | Descriptor: | GLYCEROL, SODIUM ION, Serine/threonine protein kinase | | Authors: | Burda, P.C, Crosskey, T.D, Lauk, K, Wilmanns, M, Gilberger, T.W. | | Deposit date: | 2019-12-02 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | SOLUTION SCATTERING (2.85 Å), X-RAY DIFFRACTION | | Cite: | Structure-Based Identification and Functional Characterization of a Lipocalin in the Malaria Parasite Plasmodium falciparum.
Cell Rep, 31, 2020
|
|
2RU4
 
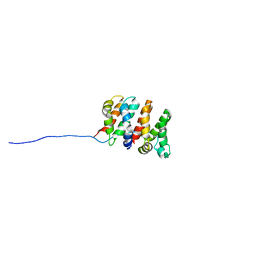 | | Designed Armadillo Repeat Protein Self-ASsembled Complex (YIIM2-MAII) | | Descriptor: | Armadillo Repeat Protein, C-terminal fragment, MAII, ... | | Authors: | Zerbe, O, Christen, M.T, Plueckthun, A, Watson, R.P. | | Deposit date: | 2013-11-22 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spontaneous self-assembly of engineered armadillo repeat protein fragments into a folded structure
Structure, 22, 2014
|
|
2RU5
 
 | |
1W0Y
 
 | | tf7a_3771 complex | | Descriptor: | 4-(4-BENZYLOXY-2-METHANESULFONYLAMINO-5-METHOXY-BENZYLAMINO)-BENZAMIDINE, BLOOD COAGULATION FACTOR VIIA, CACODYLATE ION, ... | | Authors: | Banner, D.W, D'Arcy, A, Groebke-Zbinden, K, Ackermann, J, Kirchhofer, D, Ji, Y.-H, Tschopp, T.B, Wallbaum, S, Weber, L. | | Deposit date: | 2004-05-27 | | Release date: | 2005-01-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design of Selective Phenylglycine Amide Tissue Factor/Factor Viia Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
7N6P
 
 | | Crystal structure of the anti-EBOV and SUDV monoclonal antibody 1C3 Fab | | Descriptor: | 1C3 Fab heavy chain, 1C3 Fab light chain | | Authors: | Milligan, J.C, Yu, X, Buck, T, Saphire, E.O. | | Deposit date: | 2021-06-08 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Asymmetric and non-stoichiometric glycoprotein recognition by two distinct antibodies results in broad protection against ebolaviruses.
Cell, 185, 2022
|
|
3KCU
 
 | | Structure of formate channel | | Descriptor: | 2-(6-(2-CYCLOHEXYLETHOXY)-TETRAHYDRO-4,5-DIHYDROXY-2(HYDROXYMETHYL)-2H-PYRAN-3-YLOXY)-TETRAHYDRO-6(HYDROXYMETHYL)-2H-PY RAN-3,4,5-TRIOL, Probable formate transporter 1 | | Authors: | Wang, Y, Huang, Y, Wang, J, Yan, N, Shi, Y. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel
Nature, 462, 2009
|
|
3KCV
 
 | | Structure of formate channel | | Descriptor: | Probable formate transporter 1 | | Authors: | Wang, Y, Huang, Y, Wang, J, Yan, N, Shi, Y. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | Structure of the formate transporter FocA reveals a pentameric aquaporin-like channel
Nature, 462, 2009
|
|
6J2M
 
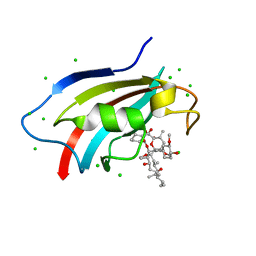 | | Crystal structure of AtFKBP53 C-terminal domain | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase FKBP53 | | Authors: | Singh, A.K, Vasudevan, D. | | Deposit date: | 2019-01-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | AtFKBP53: a chimeric histone chaperone with functional nucleoplasmin and PPIase domains.
Nucleic Acids Res., 48, 2020
|
|
6J2Z
 
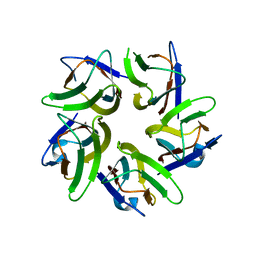 | | AtFKBP53 N-terminal Nucleoplasmin Domain | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP53 | | Authors: | Singh, A.K, Vasudevan, D. | | Deposit date: | 2019-01-03 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AtFKBP53: a chimeric histone chaperone with functional nucleoplasmin and PPIase domains.
Nucleic Acids Res., 48, 2020
|
|
3PLC
 
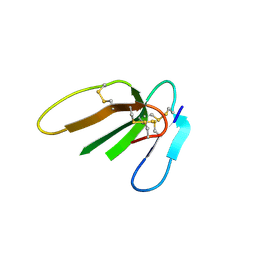 | | Crystal structure of Beta-Cardiotoxin, a novel three-finger cardiotoxin from the venom of Ophiophagus hannah | | Descriptor: | Beta-cardiotoxin OH-27 | | Authors: | Roy, A, Qingxiang, S, Kini, R.M, Sivaraman, J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-11-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Identification of a alpha-helical molten globule intermediate and structural characterization of beta-cardiotoxin, an all beta-sheet protein isolated from the venom of Ophiophagus hannah (king cobra).
Protein Sci., 28, 2019
|
|
5T6Y
 
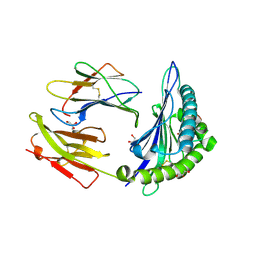 | | HLA-B*57:01 presenting TSTFEDVKILAF | | Descriptor: | ACETATE ION, Beta-2-microglobulin, Decapeptide: THR-SER-THR-PHE-GLU-ASP-VAL-LYS-ILE-LEU-ALA-PHE, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5T6Z
 
 | | KIR3DL1 in complex with HLA-B*57:01-TW10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Decapeptide: THR-SER-THR-LEU-GLN-GLU-GLN-ILE-GLY-TRP, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5T6W
 
 | | HLA-B*57:01 presenting SSTRGISQLW | | Descriptor: | Beta-2-microglobulin, Decapeptide: SER-SER-THR-ARG-GLY-ILE-SER-GLN-LEU-TRP, GLYCEROL, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-01 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5T70
 
 | | KIR3DL1 in complex with HLA-B*57:01 presenting TSNLQEQIGW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Decapeptide: THR-SER-ASN-LEU-GLN-GLU-GLN-ILE-GLY-TRP, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5T6X
 
 | | HLA-B*57:01 presenting TSTTSVASSW | | Descriptor: | Beta-2-microglobulin, Decapeptide: THR-SER-THR-THR-SER-VAL-ALA-SER-SER-TRP, GLYCEROL, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-02 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7EQX
 
 | | Crystal structure of an Aedes aegypti procarboxypeptidase B1 | | Descriptor: | Carboxypeptidase B, ZINC ION | | Authors: | Choong, Y.K, Gavor, E, Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-05-05 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of Aedes aegypti procarboxypeptidase B1 and its binding with Dengue virus for controlling infection.
Life Sci Alliance, 5, 2022
|
|
