5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
2XRO
 
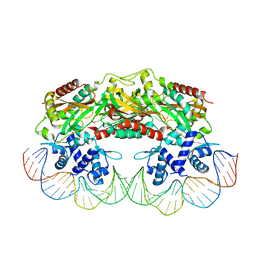 | | Crystal structure of TtgV in complex with its DNA operator | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGV, OSMIUM ION, TTGV OPERATOR DNA | | Authors: | Lu, D, Fillet, S, Meng, C, Alguel, Y, Kloppsteck, P, Bergeron, J, Krell, T, Gallegos, M.-T, Ramos, J, Zhang, X. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of Ttgv in Complex with its DNA Operator Reveals a General Model for Cooperative DNA Binding of Tetrameric Gene Regulators.
Genes Dev., 24, 2010
|
|
5XEX
 
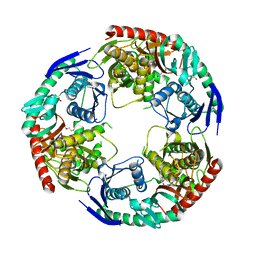 | | Crystal structure of S.aureus PNPase catalytic domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PYROPHOSPHATE, ... | | Authors: | Wang, X, Zhang, X, Zang, J. | | Deposit date: | 2017-04-06 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enolase binds to RnpA in competition with PNPase in Staphylococcus aureus
FEBS Lett., 591, 2017
|
|
2UXP
 
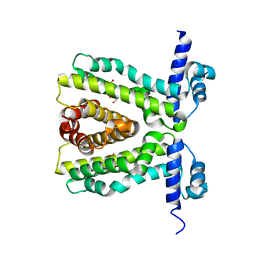 | | TtgR in complex Chloramphenicol | | Descriptor: | CHLORAMPHENICOL, HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGR | | Authors: | Alguel, Y, Meng, C, Teran, W, Krell, T, Ramos, J.L, Gallegos, M.-T, Zhang, X. | | Deposit date: | 2007-03-29 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Multidrug Binding Protein Ttgr in Complex with Antibiotics and Plant Antimicrobials.
J.Mol.Biol., 369, 2007
|
|
2UXH
 
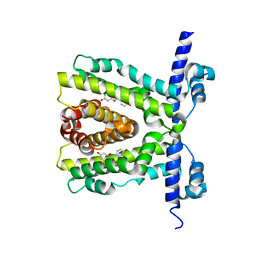 | | TtgR in complex with Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGR | | Authors: | Alguel, Y, Meng, C, Teran, W, Krell, T, Ramos, J.L, Gallegos, M.-T, Zhang, X. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Multidrug Binding Protein Ttgr in Complex with Antibiotics and Plant Antimicrobials.
J.Mol.Biol., 369, 2007
|
|
8F49
 
 | |
8F7Y
 
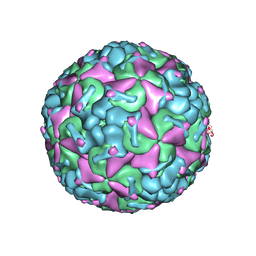 | |
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | Descriptor: | S protein | | Authors: | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | Deposit date: | 2017-02-15 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5OAF
 
 | | Human Rvb1/Rvb2 heterohexamer in INO80 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Aramayo, R.J, Bythell-Douglas, R, Ayala, R, Willhoft, O, Wigley, D, Zhang, X. | | Deposit date: | 2017-06-21 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Cryo-EM structures of the human INO80 chromatin-remodeling complex.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6HT4
 
 | |
1I85
 
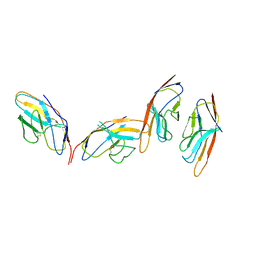 | | CRYSTAL STRUCTURE OF THE CTLA-4/B7-2 COMPLEX | | Descriptor: | CYTOTOXIC T-LYMPHOCYTE-ASSOCIATED PROTEIN 4, T LYMPHOCYTE ACTIVATION ANTIGEN CD86 | | Authors: | Schwartz, J.-C.D, Zhang, X, Fedorov, A.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2001-03-12 | | Release date: | 2001-04-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for co-stimulation by the human CTLA-4/B7-2 complex.
Nature, 410, 2001
|
|
1NGN
 
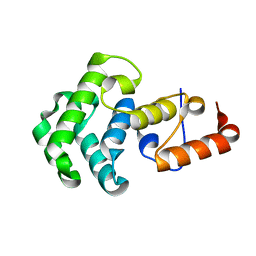 | | Mismatch repair in methylated DNA. Structure of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4 | | Descriptor: | methyl-CpG binding protein MBD4 | | Authors: | Wu, P, Qiu, C, Sohail, A, Zhang, X, Bhagwat, A.S, Cheng, X. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mismatch repair in methylated DNA. Structure and activity of the mismatch-specific thymine glycosylase domain of methyl-CpG-binding protein MBD4
J.Biol.Chem., 278, 2003
|
|
4END
 
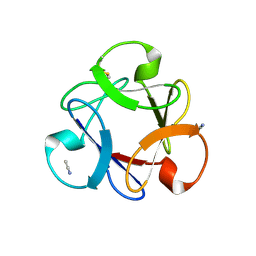 | | Crystal structure of anti-HIV actinohivin in complex with alpha-1,2-mannobiose (P 2 21 21 form) | | Descriptor: | ACETONITRILE, Actinohivin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Hoque, M.M, Suzuki, K, Tsunoda, M, Jiang, J, Zhang, F, Takahashi, A, Naomi, O, Zhang, X, Sekiguchi, T, Tanaka, H, Omura, S, Takenaka, A. | | Deposit date: | 2012-04-13 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Matured structure of anti-HIV lectin actinohivin in complex with 1,2-mannobiose
To be Published
|
|
1ICC
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5 OUTER MITOCHONDRIAL MEMBRANE ISOFORM, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Terzyan, S, Zhang, X. | | Deposit date: | 2001-03-30 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the differences between rat liver outer mitochondrial membrane cytochrome b5 and microsomal cytochromes b5.
Biochemistry, 40, 2001
|
|
8EW2
 
 | |
9JF4
 
 | | cryo-EM structure of Neuromedin B receptor in complex with PD168368 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-methyl-2-[(4-nitrophenyl)carbamoylamino]-~{N}-[(1-pyridin-2-ylcyclohexyl)methyl]propanamide, Neuromedin-B receptor,de novo design protein | | Authors: | Gao, K, Zhang, X, Liu, X. | | Deposit date: | 2024-09-03 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | cryo-EM structure of Neuromedin B receptor in complex with PD168368
To Be Published
|
|
8EW0
 
 | |
2VII
 
 | | PspF1-275-Mg-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Joly, N, Rappas, M, Buck, M, Zhang, X. | | Deposit date: | 2007-12-04 | | Release date: | 2008-01-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Trapping of a Transcription Complex Using a New Nucleotide Analogue: AMP Aluminium Fluoride
J.Mol.Biol., 375, 2008
|
|
4TOR
 
 | | Crystal structure of Tankyrase 1 with IWR-8 | | Descriptor: | 1-[(1-acetyl-5-bromo-1H-indol-6-yl)sulfonyl]-N-ethyl-N-(3-methylphenyl)piperidine-4-carboxamide, CHLORIDE ION, Tankyrase-1, ... | | Authors: | Chen, H, Zhang, X, Lum, L, Chen, C. | | Deposit date: | 2014-06-06 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Disruption of Wnt/ beta-Catenin Signaling and Telomeric Shortening Are Inextricable Consequences of Tankyrase Inhibition in Human Cells.
Mol.Cell.Biol., 35, 2015
|
|
1AWP
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, X, Zhang, X. | | Deposit date: | 1997-10-03 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The reduction potential of cytochrome b5 is modulated by its exposed heme edge.
Biochemistry, 37, 1998
|
|
8WJL
 
 | | Cryo-EM structure of 6-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.15 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8WJN
 
 | | Cryo-EM structure of 6-subunit Smc5/6 head region | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosomes element 1, Non-structural maintenance of chromosomes element 4, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (5.58 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8WJO
 
 | | Cryo-EM structure of 8-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Li, Q, Zhang, J, Zhang, X, Cheng, T, Wang, Z, Jin, D, Chen, Z, Wang, L. | | Deposit date: | 2023-09-26 | | Release date: | 2024-06-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.04 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8IQ6
 
 | | Cryo-EM structure of Latanoprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
8IQ4
 
 | | Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
