7BZ8
 
 | | Template lasso peptide C24 mutant V3A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7BZ9
 
 | | Template lasso peptide C24 mutant I4A | | Descriptor: | lasso peptide | | Authors: | Liu, X.H, Liu, T, Ma, X.J, Yu, J.H, Yang, D.H, Ma, M. | | Deposit date: | 2020-04-27 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications.
Chem Sci, 12, 2021
|
|
7DF4
 
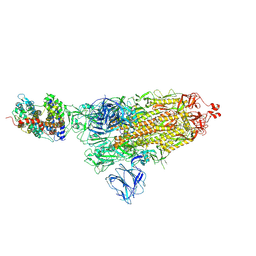 | | SARS-CoV-2 S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Cong, X, Yao, C. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7DF3
 
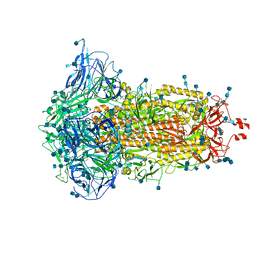 | | SARS-CoV-2 S trimer, S-closed | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cong, X, Yao, C. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7C8W
 
 | | Structure of sybody MR17 in complex with the SARS-CoV-2 S receptor-binding domain (RBD) | | Descriptor: | GLYCEROL, Spike protein S1, Synthetic nanobody MR17, ... | | Authors: | Li, T, Cai, H, Yao, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7C8V
 
 | | Structure of sybody SR4 in complex with the SARS-CoV-2 S Receptor Binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
7CAN
 
 | | Structure of sybody MR17-K99Y in complex with the SARS-CoV-2 S Receptor-binding domain (RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Spike protein S1, ... | | Authors: | Li, T, Yao, H, Cai, H, Qin, W, Li, D. | | Deposit date: | 2020-06-09 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | A synthetic nanobody targeting RBD protects hamsters from SARS-CoV-2 infection.
Nat Commun, 12, 2021
|
|
6KSB
 
 | |
6L8Q
 
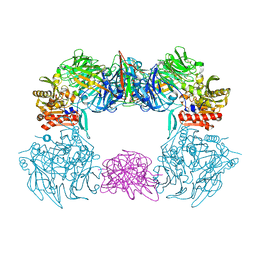 | | Complex structure of bat CD26 and MERS-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Yuan, Y. | | Deposit date: | 2019-11-07 | | Release date: | 2019-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular Basis of Binding between Middle East Respiratory Syndrome Coronavirus and CD26 from Seven Bat Species.
J.Virol., 94, 2020
|
|
6KW4
 
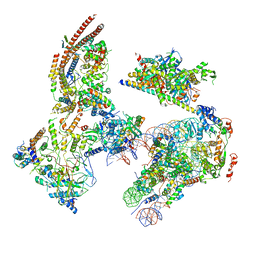 | | The ClassB RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-06 | | Release date: | 2019-11-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (7.55 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
6LQ3
 
 | |
6LQ7
 
 | |
6LQ8
 
 | |
6LPY
 
 | |
6LYP
 
 | | Cryo-EM structure of AtMSL1 wild type | | Descriptor: | Mechanosensitive ion channel protein 1, mitochondrial | | Authors: | Sun, L. | | Deposit date: | 2020-02-15 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Insights into a Plant Mechanosensitive Ion Channel MSL1.
Cell Rep, 30, 2020
|
|
6LQ5
 
 | |
7CYA
 
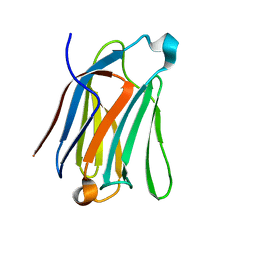 | | Saimiri boliviensis boliviensis galectin-13 with lactose | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Actin binding to galectin-13/placental protein-13 occurs independently of the galectin canonical ligand-binding site.
Glycobiology, 31, 2021
|
|
7CYB
 
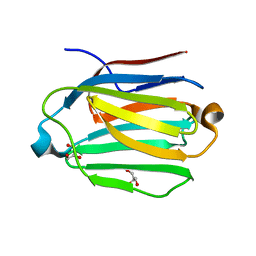 | | Saimiri boliviensis boliviensis galectin-13 with glycerol | | Descriptor: | GLYCEROL, Galectin | | Authors: | Su, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Actin binding to galectin-13/placental protein-13 occurs independently of the galectin canonical ligand-binding site.
Glycobiology, 31, 2021
|
|
7WVP
 
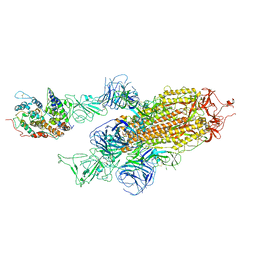 | |
7WVQ
 
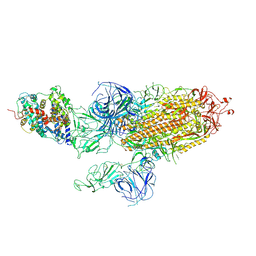 | |
3CEN
 
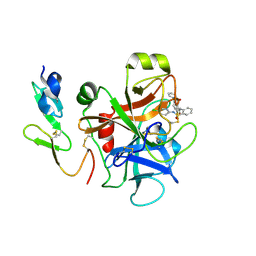 | |
1TVR
 
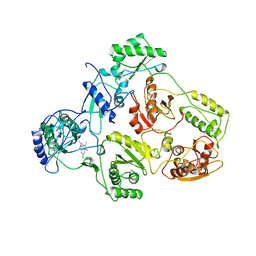 | | HIV-1 RT/9-CL TIBO | | Descriptor: | 4-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 1996-04-16 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant.
J.Mol.Biol., 264, 1996
|
|
7C02
 
 | | Crystal structure of dimeric MERS-CoV receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Dai, L, Qi, J, Gao, G.F. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS.
Cell, 182, 2020
|
|
7YR5
 
 | |
6P86
 
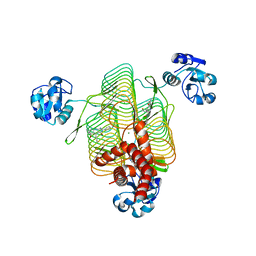 | | E.coli LpxD in complex with compound 4.1 | | Descriptor: | 3-hydroxy-7,7-dimethyl-2-phenyl-4-(thiophen-2-yl)-2,6,7,8-tetrahydro-5H-pyrazolo[3,4-b]quinolin-5-one, MAGNESIUM ION, N-[(4-ethylphenyl)methyl]-2-[(6aR)-6-oxo-3-(pyrrolidine-1-carbonyl)-6,6a,7,8,9,10-hexahydro-5H-pyrido[1,2-a]quinoxalin-5-yl]acetamide, ... | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2019-06-06 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biological Basis of Small Molecule Inhibition ofEscherichia coliLpxD Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
Acs Infect Dis., 6, 2020
|
|
