6LX5
 
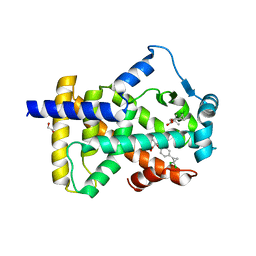 | | X-ray structure of human PPARalpha ligand binding domain-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX6
 
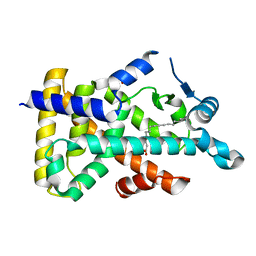 | | X-ray structure of human PPARalpha ligand binding domain-palmitic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, PALMITIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXB
 
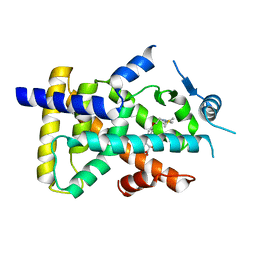 | | X-ray structure of human PPARalpha ligand binding domain-saroglitazar co-crystals obtained by soaking | | Descriptor: | (2S)-2-ethoxy-3-[4-[2-[2-methyl-5-(4-methylsulfanylphenyl)pyrrol-1-yl]ethoxy]phenyl]propanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Honda, A, Ishikawa, R, Akahane, M, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXA
 
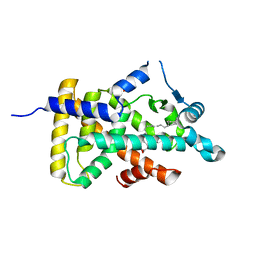 | | X-ray structure of human PPARalpha ligand binding domain-eicosapentaenoic acid (EPA) co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
8GS6
 
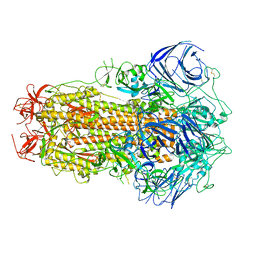 | | Structure of the SARS-CoV-2 BA.2.75 spike glycoprotein (closed state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Anraku, Y, Tabata-Sasaki, K, Kita, S, Fukuhara, H, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2022-09-05 | | Release date: | 2022-10-26 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron BA.2.75 variant.
Cell Host Microbe, 30, 2022
|
|
8JYM
 
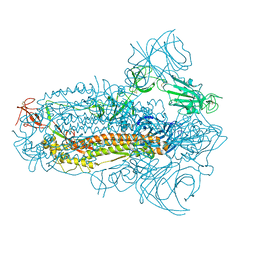 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYK
 
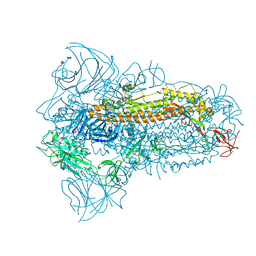 | | Structure of the SARS-CoV-2 XBB.1.5 spike glycoprotein (closed state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYP
 
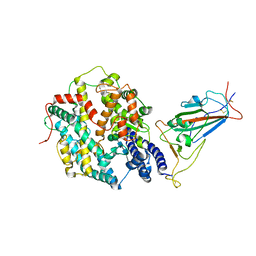 | | Structure of SARS-CoV-2 XBB.1.5 spike RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYN
 
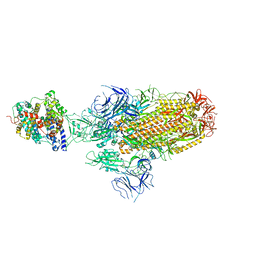 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
8JYO
 
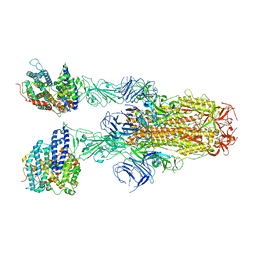 | | Structure of SARS-CoV-2 XBB.1.5 spike glycoprotein in complex with ACE2 (2-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Yajima, H, Anraku, Y, Kita, S, Kimura, K, Sasaki, J, Sasaki-Tabata, K, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2023-07-03 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 XBB.1.5 variant
To Be Published
|
|
4ZFZ
 
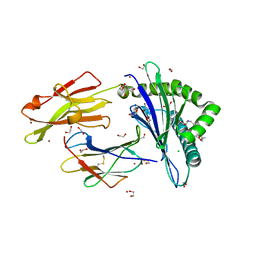 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with myristoylated 5-mer lipopeptide derived from SIV Nef protein | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-mer lipopeptide from Protein Nef, ... | | Authors: | Morita, D, Sugita, M. | | Deposit date: | 2015-04-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.763 Å) | | Cite: | Crystal structure of the N-myristoylated lipopeptide-bound MHC class I complex
Nat Commun, 7, 2016
|
|
2JMW
 
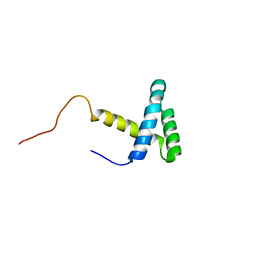 | | Structure of DNA-Binding Domain of Arabidopsis GT-1 | | Descriptor: | DNA binding protein GT-1 | | Authors: | Nagata, T, Niyada, E, Noto, K, Ikeda, Y, Yamamoto, Y, Uesugi, S, Murata, J, Hiratsuka, K, Katahira, M. | | Deposit date: | 2006-12-11 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation.
Proteins, 78, 2010
|
|
3WV2
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with N-(3-methoxybenzyl)-4-oxo-3,4-dihydroquinazoline-2-carboxamide | | Descriptor: | CALCIUM ION, Collagenase 3, N-(3-methoxybenzyl)-4-oxo-3,4-dihydroquinazoline-2-carboxamide, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thieno[2,3-d]pyrimidine-2-carboxamides bearing a carboxybenzene group at 5-position: highly potent, selective, and orally available MMP-13 inhibitors interacting with the S1′′ binding site.
Bioorg.Med.Chem., 22, 2014
|
|
3WV3
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with N-(3-methoxybenzyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2014-05-12 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Thieno[2,3-d]pyrimidine-2-carboxamides bearing a carboxybenzene group at 5-position: highly potent, selective, and orally available MMP-13 inhibitors interacting with the S1′′ binding site.
Bioorg.Med.Chem., 22, 2014
|
|
3WV1
 
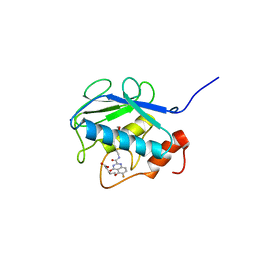 | | Crystal structure of the catalytic domain of MMP-13 complexed with 4-(2-((6-fluoro-2-((3-methoxybenzyl)carbamoyl)-4-oxo-3,4-dihydroquinazolin-5-yl)oxy)ethyl)benzoic acid | | Descriptor: | 4-[2-({6-fluoro-2-[(3-methoxybenzyl)carbamoyl]-4-oxo-3,4-dihydroquinazolin-5-yl}oxy)ethyl]benzoic acid, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2014-05-12 | | Release date: | 2015-05-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of novel, highly potent, and selective quinazoline-2-carboxamide-based matrix metalloproteinase (MMP)-13 inhibitors without a zinc binding group using a structure-based design approach
J.Med.Chem., 57, 2014
|
|
1J0S
 
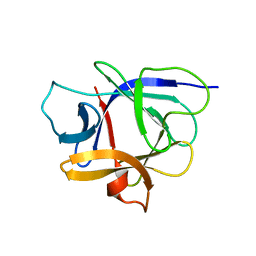 | | Solution structure of the human interleukin-18 | | Descriptor: | Interleukin-18 | | Authors: | Kato, Z, Jee, J, Shikano, H, Mishima, M, Ohki, I, Yoneda, T, Hara, T, Torigoe, K, Kondo, N, Shirakawa, M. | | Deposit date: | 2002-11-21 | | Release date: | 2003-11-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The structure and binding mode of interleukin-18
Nat.Struct.Biol., 10, 2003
|
|
3ALP
 
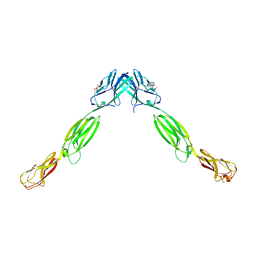 | | Cell adhesion protein | | Descriptor: | CITRIC ACID, HEXANE-1,6-DIOL, Poliovirus receptor-related protein 1 | | Authors: | Narita, H, Nakagawa, A, Suzuki, M. | | Deposit date: | 2010-08-05 | | Release date: | 2011-02-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Crystal Structure of the cis-Dimer of Nectin-1: implications for the architecture of cell-cell junctions
J.Biol.Chem., 286, 2011
|
|
1LHM
 
 | |
5B5O
 
 | | Crystal structure of the catalytic domain of MMP-13 complexed with N-phenyl-4-((4H-1,2,4-triazol-3-ylsulfanyl)methyl)-1,3-thiazol-2-amine | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2016-05-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of Novel, Highly Potent, and Selective Matrix Metalloproteinase (MMP)-13 Inhibitors with a 1,2,4-Triazol-3-yl Moiety as a Zinc Binding Group Using a Structure-Based Design Approach
J. Med. Chem., 60, 2017
|
|
5B5P
 
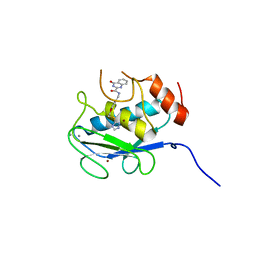 | | Crystal structure of the catalytic domain of MMP-13 complexed with 4-oxo-N-(3-(2-(1H-1,2,4-triazol-3-ylsulfanyl)ethoxy)benzyl)-3,4-dihydroquinazoline-2-carboxamide | | Descriptor: | 4-oxo-N-{3-[2-(1H-1,2,4-triazol-3-ylsulfanyl)ethoxy]benzyl}-3,4-dihydroquinazoline-2-carboxamide, CALCIUM ION, Collagenase 3, ... | | Authors: | Oki, H, Tanaka, Y. | | Deposit date: | 2016-05-13 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Novel, Highly Potent, and Selective Matrix Metalloproteinase (MMP)-13 Inhibitors with a 1,2,4-Triazol-3-yl Moiety as a Zinc Binding Group Using a Structure-Based Design Approach
J. Med. Chem., 60, 2017
|
|
5EGE
 
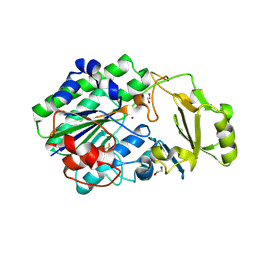 | | Structure of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Morita, J, Kano, K, Kato, K, Takita, H, Ishitani, R, Nishimasu, H, Nureki, O, Aoki, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase
Sci Rep, 6, 2016
|
|
5EGH
 
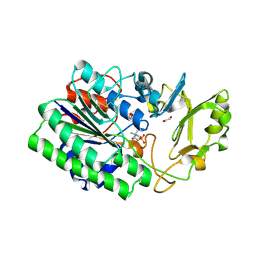 | | Structure of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase in complex with phosphocholine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Morita, J, Kano, K, Kato, K, Takita, H, Ishitani, R, Nishimasu, H, Nureki, O, Aoki, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase
Sci Rep, 6, 2016
|
|
7XWA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BA.4/5 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Suzuki, T, Kimura, K, Hashiguchi, T. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5.
Cell, 185, 2022
|
|
7VHZ
 
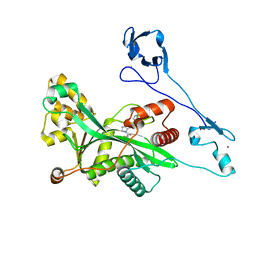 | | Crystal structure of EP300 HAT domain in complex with compound 7 | | Descriptor: | (2R)-N-(2H-indazol-4-yl)-1-[1-(4-methoxyphenyl)cyclopentyl]carbonyl-pyrrolidine-2-carboxamide, Histone acetyltransferase p300, ZINC ION | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2021-09-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of EP300/CBP histone acetyltransferase inhibitors through scaffold hopping of 1,4-oxazepane ring.
Bioorg.Med.Chem.Lett., 66, 2022
|
|
7VHY
 
 | | Crystal structure of EP300 HAT domain in complex with compound (+)-3 | | Descriptor: | Histone acetyltransferase p300, ZINC ION, [(6R)-6-(1H-indazol-4-ylmethyl)-1,4-oxazepan-4-yl]-[1-(4-methoxyphenyl)cyclopentyl]methanone | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2021-09-24 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of EP300/CBP histone acetyltransferase inhibitors through scaffold hopping of 1,4-oxazepane ring.
Bioorg.Med.Chem.Lett., 66, 2022
|
|
