6A3K
 
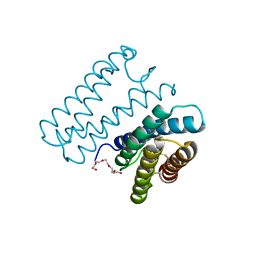 | | Crystal structure of cytochrome c' from Shewanella benthica DB6705 | | Descriptor: | Cytochrome c, HEME C, PENTAETHYLENE GLYCOL | | Authors: | Suka, A, Oki, H, Kato, Y, Kawahara, K, Ohkubo, T, Maruno, T, Kobayashi, Y, Fujii, S, Wakai, S, Sambongi, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-12 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Stability of cytochromes c' from psychrophilic and piezophilic Shewanella species: implications for complex multiple adaptation to low temperature and high hydrostatic pressure.
Extremophiles, 23, 2019
|
|
1IRZ
 
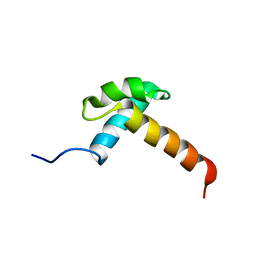 | |
1HNR
 
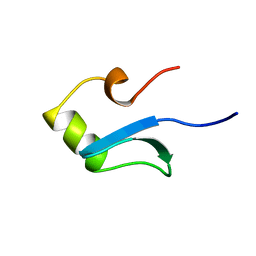 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
1HNS
 
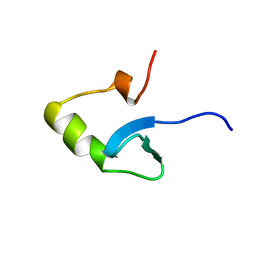 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
2RQQ
 
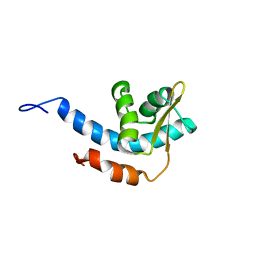 | | Structure of C-terminal region of Cdt1 | | Descriptor: | DNA replication factor Cdt1 | | Authors: | Jee, J.G, Mizuno, T, Kamada, K, Tochio, H, Hiroaki, H, Hanaoka, F, Shirakawa, M. | | Deposit date: | 2009-10-14 | | Release date: | 2010-03-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and mutagenesis studies of the C-terminal region of licensing factor Cdt1 enable the identification of key residues for binding to replicative helicase Mcm proteins
J.Biol.Chem., 285, 2010
|
|
4Z55
 
 | |
4PX6
 
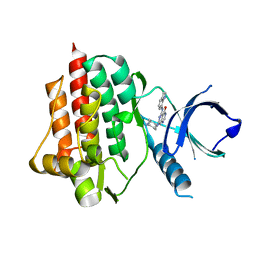 | | SYK catalytic domain in complex with a potent pyridopyrimidinone inhibitor | | Descriptor: | 7-{[(1R,2S)-2-aminocyclohexyl]amino}-5-(1H-indol-7-ylamino)pyrido[4,3-d]pyrimidin-4(3H)-one, Tyrosine-protein kinase SYK | | Authors: | Lee, C.C. | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Syk inhibitors with high potency in presence of blood.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6BI2
 
 | |
6BHZ
 
 | | Trastuzumab Fab D185A (Light Chain) Mutant. | | Descriptor: | 1,2-ETHANEDIOL, Trastuzumab Anti-HER2 Fab Heavy Chain, Trastuzumab Anti-HER2 Fab Light Chain D185A | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Tuning a Protein-Labeling Reaction to Achieve Highly Site Selective Lysine Conjugation.
Chembiochem, 19, 2018
|
|
6BI0
 
 | | Trastuzumab Fab N158A, D185A, K190A (Light Chain) Triple Mutant. | | Descriptor: | 1,2-ETHANEDIOL, Trastuzumab anti-HER2 Fab Heavy Chain, Trastuzumab anti-HER2 Fab Light Chain | | Authors: | DiDonato, M, Spraggon, G. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.057 Å) | | Cite: | Tuning a Protein-Labeling Reaction to Achieve Highly Site Selective Lysine Conjugation.
Chembiochem, 19, 2018
|
|
4ZY3
 
 | | Crystal Structure of Keap1 in Complex with a small chemical compound, K67 | | Descriptor: | FORMIC ACID, Kelch-like ECH-associated protein 1, N,N'-[2-(2-oxopropyl)naphthalene-1,4-diyl]bis(4-ethoxybenzenesulfonamide) | | Authors: | Fukutomi, T, Iso, T, Suzuki, T, Takagi, K, Mizushima, T, Komatsu, M, Yamamoto, M. | | Deposit date: | 2015-05-21 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming
Nat Commun, 7, 2016
|
|
6LFE
 
 | | Rat-COMT, Nitecapone,SAM and Mg bond | | Descriptor: | 3-(3,4-dihydroxy-5-nitrobenzylidene)pentane-2,4-dione, Catechol O-methyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Takebe, K, Iijima, H, Suzuki, M. | | Deposit date: | 2019-12-02 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Catechol O-Methyltransferase Complexed with Nitecapone.
Chem Pharm Bull (Tokyo), 68, 2020
|
|
8K2K
 
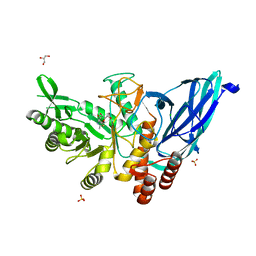 | | Crystal structure of Group 3 Oligosaccharide/Monosaccharide-releasing beta-N-acetylgalactosaminidase NgaDssm in complex with GalNAc-thiazoline | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
8K2J
 
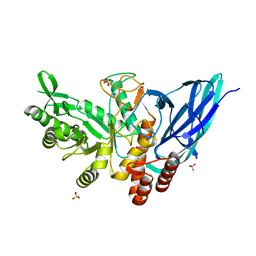 | |
8K2G
 
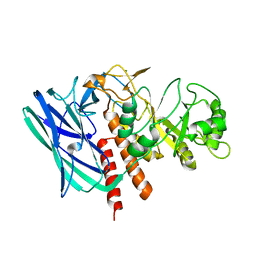 | |
8K2I
 
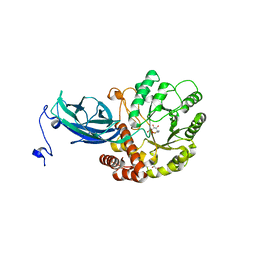 | | Crystal structure of Group 2 Oligosaccharide/Monosaccharide-releasing beta-N-acetylhexosaminidase NgaAt from Arabidopsis thaliana in complex with GlcNAc-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
8K2L
 
 | |
8K2H
 
 | | Crystal structure of Group 2Oligosaccharide/Monosaccharide-releasing beta-N-acetylhexosaminidase NgaAt from Arabidopsis thaliana in complex with GalNAc-thiazoline | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
8K2F
 
 | |
8K2M
 
 | | Crystal structure of Group 4 Monosaccharide-releasing beta-N-acetylgalactosaminidase NgaP2 from Paenibacillus sp. TS12 in complex with GalNAc-thiazoline | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, BROMIDE ION, Monosaccharide-releasing beta-N-acetylgalactosaminidase | | Authors: | Sumida, T, Fushinobu, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Genetic and functional diversity of beta-N-acetylgalactosamine-targeting glycosidases expanded by deep-sea metagenome analysis.
Nat Commun, 15, 2024
|
|
8K2N
 
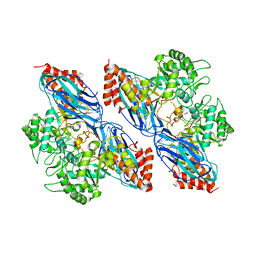 | |
5X33
 
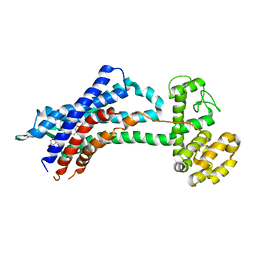 | | Leukotriene B4 receptor BLT1 in complex with BIIL260 | | Descriptor: | 4-[[3-[[4-[2-(4-hydroxyphenyl)propan-2-yl]phenoxy]methyl]phenyl]methoxy]benzenecarboximidamide, LTB4 receptor,Lysozyme,LTB4 receptor | | Authors: | Hori, T, Hirata, K, Yamashita, K, Kawano, Y, Yamamoto, M, Yokoyama, S. | | Deposit date: | 2017-02-03 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Na+-mimicking ligands stabilize the inactive state of leukotriene B4receptor BLT1.
Nat. Chem. Biol., 14, 2018
|
|
7B7V
 
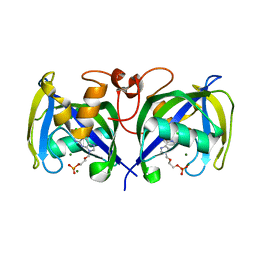 | | Structure of NUDT15 in complex with Acyclovir monophosphate | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl dihydrogen phosphate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NUDT15 polymorphism influences the metabolism and therapeutic effects of acyclovir and ganciclovir.
Nat Commun, 12, 2021
|
|
7BTV
 
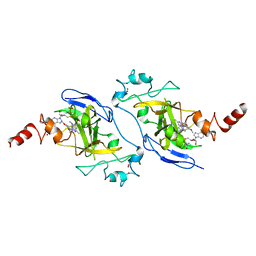 | | Crystal structure of EHMT2 SET domain in complex with compound 5. | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N~2~-{4-methoxy-3-[3-(pyrrolidin-1-yl)propoxy]phenyl}-N~4~,6-dimethylpyrimidine-2,4-diamine, S-ADENOSYLMETHIONINE, ... | | Authors: | Suzuki, M, Mizuno, T, Katayama, K. | | Deposit date: | 2020-04-03 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel histone lysine methyltransferase G9a/GLP (EHMT2/1) inhibitors: Design, synthesis, and structure-activity relationships of 2,4-diamino-6-methylpyrimidines.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
3W8K
 
 | | Crystal structure of class C beta-lactamase Mox-1 | | Descriptor: | ACETATE ION, Beta-lactamase, ZINC ION | | Authors: | Shimizu-ibuka, A, Oguri, T, Furuyama, T, Ishii, Y. | | Deposit date: | 2013-03-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Mox-1, a unique plasmid-mediated class C beta-lactamase with hydrolytic activity towards moxalactam
Antimicrob.Agents Chemother., 58, 2014
|
|
