2QG2
 
 | | HSP90 complexed with A917985 | | Descriptor: | 3-({2-[(2-AMINO-6-METHYLPYRIMIDIN-4-YL)ETHYNYL]BENZYL}AMINO)-1,3-OXAZOL-2(3H)-ONE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QG0
 
 | | HSP90 complexed with A943037 | | Descriptor: | Heat shock protein HSP 90-alpha, N-[(2-AMINO-6-METHYLPYRIMIDIN-4-YL)METHYL]-3-{[(E)-(2-OXODIHYDROFURAN-3(2H)-YLIDENE)METHYL]AMINO}BENZENESULFONAMIDE | | Authors: | Park, C.H. | | Deposit date: | 2007-06-28 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
Chem.Biol.Drug Des., 70, 2007
|
|
2QFO
 
 | | HSP90 complexed with A143571 and A516383 | | Descriptor: | (3E)-3-[(phenylamino)methylidene]dihydrofuran-2(3H)-one, 4-METHYL-6-(TRIFLUOROMETHYL)PYRIMIDIN-2-AMINE, Heat shock protein HSP 90-alpha | | Authors: | Park, C.H. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery and design of novel HSP90 inhibitors using multiple fragment-based design strategies.
CHEM.BIOL.DRUG DES., 70, 2007
|
|
2EA2
 
 | | h-MetAP2 complexed with A773812 | | Descriptor: | 3-ETHYL-6-{[(4-FLUOROPHENYL)SULFONYL]AMINO}-2-METHYLBENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lead optimization of methionine aminopeptidase-2 (MetAP2) inhibitors containing sulfonamides of 5,6-disubstituted anthranilic acids
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2EA4
 
 | | h-MetAP2 complexed with A797859 | | Descriptor: | 2-(2-AMINOETHOXY)-3-ETHYL-6-{[(4-FLUOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C.H. | | Deposit date: | 2007-01-30 | | Release date: | 2008-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Lead optimization of methionine aminopeptidase-2 (MetAP2) inhibitors containing sulfonamides of 5,6-disubstituted anthranilic acids
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2PF2
 
 | | THE CA+2 ION AND MEMBRANE BINDING STRUCTURE OF THE GLA DOMAIN OF CA-PROTHROMBIN FRAGMENT 1 | | Descriptor: | CALCIUM ION, PROTHROMBIN FRAGMENT 1 | | Authors: | Soriano-Garcia, M, Padmanabhan, K, De vos, A.M, Tulinsky, A. | | Deposit date: | 1991-12-08 | | Release date: | 1994-01-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Ca2+ ion and membrane binding structure of the Gla domain of Ca-prothrombin fragment 1.
Biochemistry, 31, 1992
|
|
8GPZ
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with C239-0012 | | Descriptor: | 3-methyl-6-(4-methylpiperidin-1-yl)-[1,2,4]triazolo[4,3-b]pyridazine, Bromodomain-containing protein 4, FORMIC ACID, ... | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2022-08-27 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.528 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
8GQ0
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with STL233497 | | Descriptor: | Bromodomain-containing protein 4, FORMIC ACID, GLYCEROL, ... | | Authors: | Park, T.H, Lee, B.I. | | Deposit date: | 2022-08-27 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
8WPL
 
 | | Cryo-EM structure of the human TRPC1/C4 heteromer | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Cryo-EM structure of the heteromeric TRPC1/TRPC4 channel.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8WPM
 
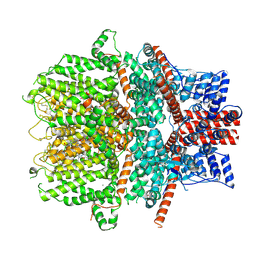 | | Cryo-EM structure of the human TRPC1/C4 heteromer in complex with Pico145 | | Descriptor: | 7-[(4-chlorophenyl)methyl]-3-methyl-1-(3-oxidanylpropyl)-8-[3-(trifluoromethyloxy)phenoxy]purine-2,6-dione, CALCIUM ION, Short transient receptor potential channel 1, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Cryo-EM structure of the heteromeric TRPC1/TRPC4 channel.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8WPN
 
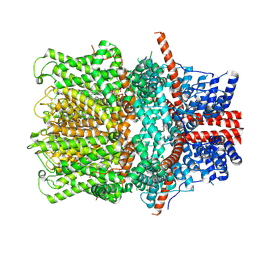 | | Cryo-EM structure of the human TRPC4 in lipid nanodiscs | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Won, J, Jeong, H, Lee, H.H. | | Deposit date: | 2023-10-10 | | Release date: | 2024-10-16 | | Last modified: | 2025-03-05 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Cryo-EM structure of the heteromeric TRPC1/TRPC4 channel.
Nat.Struct.Mol.Biol., 32, 2025
|
|
1PK4
 
 | |
4AEE
 
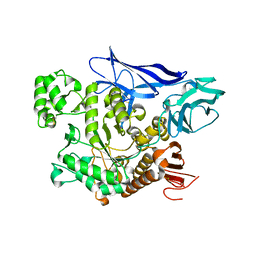 | | CRYSTAL STRUCTURE OF MALTOGENIC AMYLASE FROM S.MARINUS | | Descriptor: | ALPHA AMYLASE, CATALYTIC REGION | | Authors: | Jung, T.Y, Park, C.H, Yoon, S.M, Park, S.H, Park, K.H, Woo, E.J. | | Deposit date: | 2012-01-10 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Association of Novel Domain in Active Site of Archaic Hyperthermophilic Maltogenic Amylase from Staphylothermus Marinus.
J.Biol.Chem., 287, 2012
|
|
7C0J
 
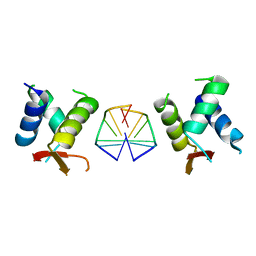 | | Crystal structure of chimeric mutant of GH5 in complex with Z-DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), Histone H5,Double-stranded RNA-specific adenosine deaminase | | Authors: | Choi, H.J, Park, C.H. | | Deposit date: | 2020-05-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process.
Nucleic Acids Res., 48, 2020
|
|
7C0I
 
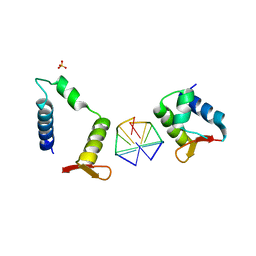 | | Crystal structure of chimeric mutant of E3L in complex with Z-DNA | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), Double-stranded RNA-binding protein,Double-stranded RNA-specific adenosine deaminase, SULFATE ION | | Authors: | Choi, H.J, Park, C.H, Kim, J.S. | | Deposit date: | 2020-05-01 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dual conformational recognition by Z-DNA binding protein is important for the B-Z transition process.
Nucleic Acids Res., 48, 2020
|
|
7YMG
 
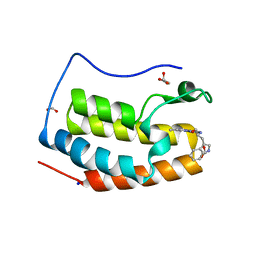 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 2-({3-ethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl}amino)-3-(1H-indol-3-yl)propan-1-ol | | Descriptor: | (2S)-2-[(3-ethyl-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)amino]-3-(1H-indol-3-yl)propan-1-ol, Bromodomain-containing protein 4, FORMIC ACID, ... | | Authors: | Kim, J.H, Lee, B.I. | | Deposit date: | 2022-07-28 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
7YQ9
 
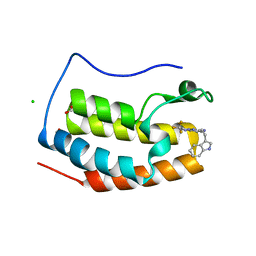 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with N-[2-(1H-indol-3-yl)ethyl]-3-(trifluoromethyl)[1,2,4]triazolo[4,3-b]pyridazin-6-amine | | Descriptor: | Bromodomain-containing protein 4, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, J.H, Lee, B.I. | | Deposit date: | 2022-08-05 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of [1,2,4]triazolo[4,3-b]pyridazine derivatives as BRD4 bromodomain inhibitors and structure-activity relationship study.
Sci Rep, 13, 2023
|
|
6VWC
 
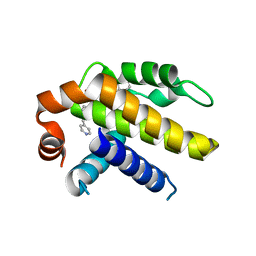 | | Crystal structure of Bcl-xL in complex with tetrahydroisoquinoline-pyridine based inhibitors | | Descriptor: | 6-{8-[(1,3-benzothiazol-2-yl)carbamoyl]-3,4-dihydroisoquinolin-2(1H)-yl}-3-{1-[(pyridin-4-yl)methyl]-1H-pyrazol-4-yl}pyridine-2-carboxylic acid, Bcl-2-like protein 1 | | Authors: | Judge, R.A, Judd, A.S. | | Deposit date: | 2020-02-19 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Discovery of A-1331852, a First-in-Class, Potent, and Orally-Bioavailable BCL-X L Inhibitor.
Acs Med.Chem.Lett., 11, 2020
|
|
6ONY
 
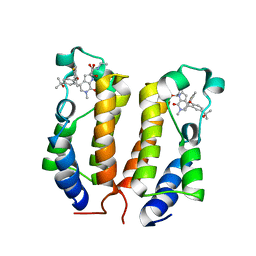 | | BRD2_Bromodomain1 complex with inhibitor 744 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, N-ethyl-4-[2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl]-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Longenecker, K.L, Bigelow, L. | | Deposit date: | 2019-04-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer.
Nature, 578, 2020
|
|
6KEI
 
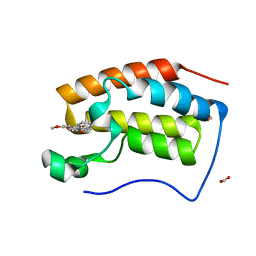 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 16-methoxy-11-methyl-6-[(pyridin-2-yl)methoxy]-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one | | Descriptor: | 16-methoxy-11-methyl-6-[(pyridin-2-yl)methoxy]-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
6KEC
 
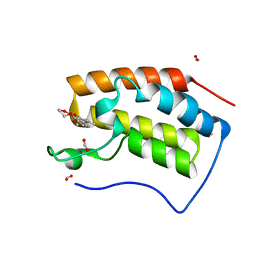 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 4-ethoxy-5,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one | | Descriptor: | 4-ethoxy-5,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
6KEH
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 6,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one | | Descriptor: | 6,16-dimethoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
6KEJ
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 6-[2-(diethylamino)ethoxy]-16-methoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one | | Descriptor: | 6-[2-(diethylamino)ethoxy]-16-methoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(17),3,5,7,9,13,15-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
2PK4
 
 | |
6KEK
 
 | | Crystal structure of BRD4 bromodomain 1 (BD1) in complex with 6-hydroxy-16-methoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one | | Descriptor: | 6-hydroxy-16-methoxy-11-methyl-2-oxa-11-azatetracyclo[8.6.1.03,8.013,17]heptadeca-1(16),3,5,7,9,13(17),14-heptaen-12-one, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Lee, B.I, Park, T.H. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Synthesis and Structure-Activity Relationships of Aristoyagonine Derivatives as Brd4 Bromodomain Inhibitors with X-ray Co-Crystal Research.
Molecules, 26, 2021
|
|
