2D1T
 
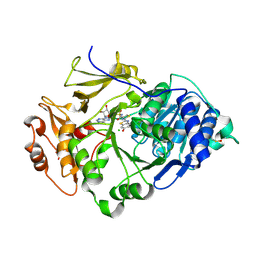 | | Crystal structure of the thermostable Japanese Firefly Luciferase red-color emission S286N mutant complexed with High-energy intermediate analogue | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
7DQV
 
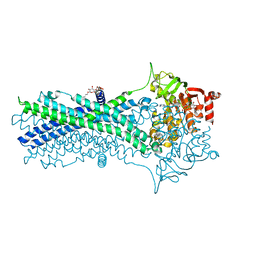 | | Crystal structure of a CmABCB1 mutant | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Matsuoka, K, Nakatsu, T, Kato, H. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the CmABCB1 G132V mutant, which favors the outward-facing state, reveals the mechanism of the pivotal joint between TM1 and TM3.
Protein Sci., 30, 2021
|
|
3W15
 
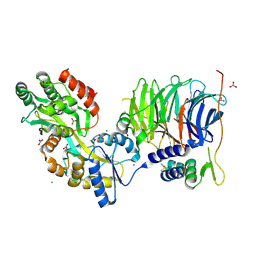 | | Structure of peroxisomal targeting signal 2 (PTS2) of Saccharomyces cerevisiae 3-ketoacyl-CoA thiolase in complex with Pex7p and Pex21p | | Descriptor: | 3-ketoacyl-CoA thiolase, peroxisomal, Maltose-binding periplasmic protein, ... | | Authors: | Pan, D, Nakatsu, T, Kato, H. | | Deposit date: | 2012-11-06 | | Release date: | 2013-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p
Nat.Struct.Mol.Biol., 20, 2013
|
|
2K6Q
 
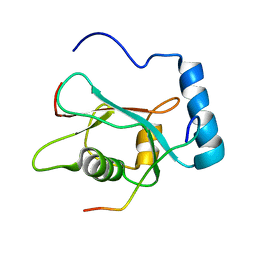 | | LC3 p62 complex structure | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, p62_peptide from Sequestosome-1 | | Authors: | Noda, N, Kumeta, H, Nakatogawa, H, Satoo, K, Adachi, W, Ishii, J, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2008-07-17 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of target recognition by ATG8/LC3 during selective autophagy
To be Published
|
|
2KWC
 
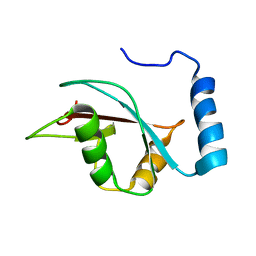 | | The NMR structure of the autophagy-related protein Atg8 | | Descriptor: | Autophagy-related protein 8 | | Authors: | Kumeta, H, Watanabe, M, Nakatogawa, H, Yamaguchi, M, Ogura, K, Adachi, W, Fujioka, Y, Noda, N.N, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2010-04-05 | | Release date: | 2010-05-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of the autophagy-related protein Atg8
J.Biomol.Nmr, 47, 2010
|
|
2LI5
 
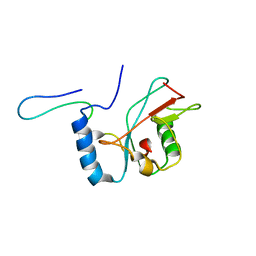 | | NMR structure of Atg8-Atg7C30 complex | | Descriptor: | Autophagy-related protein 8, Ubiquitin-like modifier-activating enzyme ATG7 | | Authors: | Kumeta, H, Satoo, K, Noda, N.N, Fujioka, Y, Ogura, K, Nakatogawa, H, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2011-08-23 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of Atg8 activation by a homodimeric E1, Atg7.
Mol.Cell, 44, 2011
|
|
3VX7
 
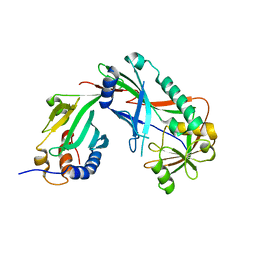 | | Crystal structure of Kluyveromyces marxianus Atg7NTD-Atg10 complex | | Descriptor: | E1, E2 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VX6
 
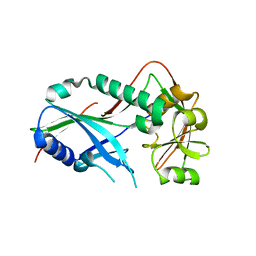 | | Crystal structure of Kluyveromyces marxianus Atg7NTD | | Descriptor: | E1 | | Authors: | Yamaguchi, M, Matoba, K, Sawada, R, Fujioka, Y, Nakatogawa, H, Yamamoto, H, Kobashigawa, Y, Hoshida, H, Akada, R, Ohsumi, Y, Noda, N.N, Inagaki, F. | | Deposit date: | 2012-09-11 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6SDG
 
 | |
4X23
 
 | |
5XFC
 
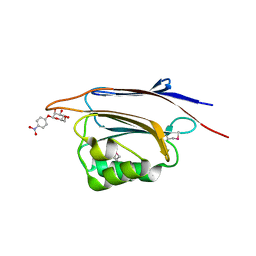 | | Serial femtosecond X-ray structure of a stem domain of human O-mannose beta-1,2-N-acetylglucosaminyltransferase solved by Se-SAD using XFEL (refined against 13,000 patterns) | | Descriptor: | 4-nitrophenyl beta-D-mannopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Fumiaki, Y, Kato, R, Manya, H. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental phase determination with selenomethionine or mercury-derivatization in serial femtosecond crystallography
IUCrJ, 4, 2017
|
|
5XFD
 
 | |
1IQW
 
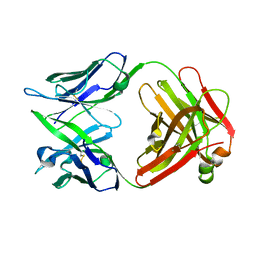 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF THE MOUSE ANTI-HUMAN FAS ANTIBODY HFE7A | | Descriptor: | ANTIBODY M-HFE7A, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Ito, S, Takayama, T, Hanzawa, H, Ichikawa, K, Ohsumi, J, Serizawa, N, Hata, T, Haruyama, H. | | Deposit date: | 2001-08-10 | | Release date: | 2002-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the antigen-binding fragment of apoptosis-inducing mouse anti-human Fas monoclonal antibody HFE7A.
J.Biochem., 131, 2002
|
|
7B17
 
 | | SARS-CoV-spike RBD bound to two neutralising nanobodies. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SARS-CoV-2 neutralizing biparatopic nanobody VE,nanobody E from Lama glama,SARS-CoV-2 neutralizing biparatopic nanobody VE,nanobody E from Lama glama, Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Science, 371, 2021
|
|
7B14
 
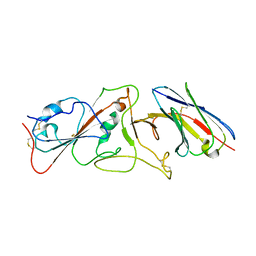 | | Nanobody E bound to Spike-RBD in a localized reconstruction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2, Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-23 | | Release date: | 2021-04-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Science, 371, 2021
|
|
7B18
 
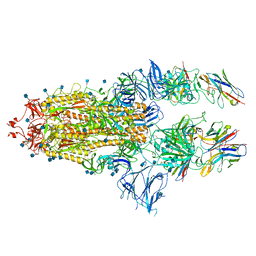 | | SARS-CoV-spike bound to two neutralising nanobodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 VHH E, ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7KN6
 
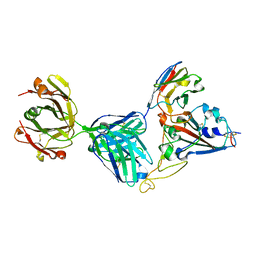 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobody VHH V and antibody Fab CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, CC12.3 Fab light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7KN7
 
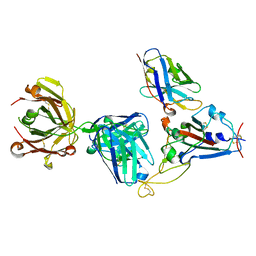 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobody VHH W and antibody Fab CC12.3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CC12.3 Fab heavy chain, CC12.3 Fab light chain, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
1IT9
 
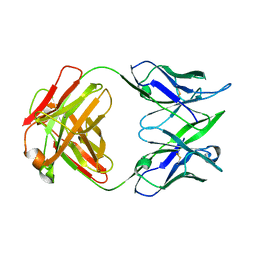 | | CRYSTAL STRUCTURE OF AN ANTIGEN-BINDING FRAGMENT FROM A HUMANIZED VERSION OF THE ANTI-HUMAN FAS ANTIBODY HFE7A | | Descriptor: | HUMANIZED ANTIBODY HFE7A, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Ito, S, Takayama, T, Hanzawa, H, Takahashi, T, Miyadai, K, Serizawa, N, Hata, T, Haruyama, H. | | Deposit date: | 2002-01-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Humanization of the Mouse Anti-Fas Antibody HFE7A and Crystal Structure of the Humanized HFE7A Fab Fragment
BIOL.PHARM.BULL., 25, 2002
|
|
7KN5
 
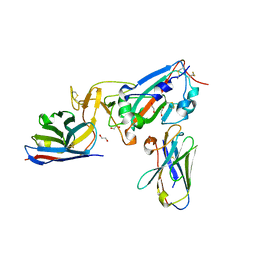 | | Crystal structure of SARS-CoV-2 receptor binding domain complexed with nanobodies VHH E and U | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, H, Yuan, M, Zhu, X, Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7KSG
 
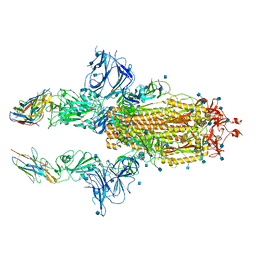 | | SARS-CoV-2 spike in complex with nanobodies E | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 glycoprotein, Spike glycoprotein | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-22 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
5XEO
 
 | |
5XEN
 
 | |
5H2B
 
 | | Structure of a novel antibody G196 | | Descriptor: | G196 antibody Heavy chain, G196 antibody Light chain | | Authors: | Park, S.Y, Sugiyama, K. | | Deposit date: | 2016-10-14 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | G196 epitope tag system: a novel monoclonal antibody, G196, recognizes the small, soluble peptide DLVPR with high affinity.
Sci Rep, 7, 2017
|
|
7XMC
 
 | | Cryo-EM structure of Cytochrome bo3 from Escherichia coli, apo structure with DMSO | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Nishida, Y, Shigematsu, H, Iwamoto, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
