4RHS
 
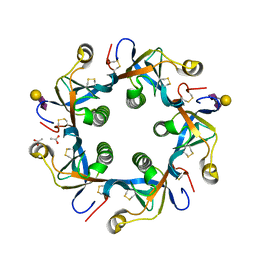 | | Crystal structure of GD2 bound PltB | | Descriptor: | ACETATE ION, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, Putative pertussis-like toxin subunit | | Authors: | Gao, X, Wang, J, Galan, J. | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9211 Å) | | Cite: | Host adaptation of a bacterial toxin from the human pathogen salmonella typhi.
Cell(Cambridge,Mass.), 159, 2014
|
|
4LCL
 
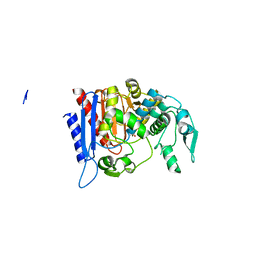 | | Simvastatin Synthase (LOVD), from Aspergillus Terreus, LovD6 mutant (simh6208) | | Descriptor: | ISOPROPYL ALCOHOL, Transesterase | | Authors: | Gao, X, Sawaya, M.R, Yeates, T.O, Tang, Y. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The role of distant mutations and allosteric regulation on LovD active site dynamics.
Nat.Chem.Biol., 10, 2014
|
|
4LCM
 
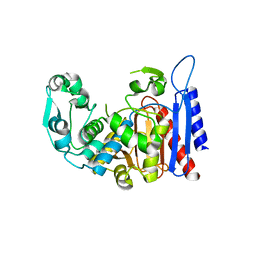 | | Simvastatin Synthase (LOVD), from Aspergillus Terreus, LovD9 mutant (simh9014) | | Descriptor: | Transesterase | | Authors: | Gao, X, Sawaya, M.R, Yeates, T.O, Tang, Y. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The role of distant mutations and allosteric regulation on LovD active site dynamics.
Nat.Chem.Biol., 10, 2014
|
|
4K6L
 
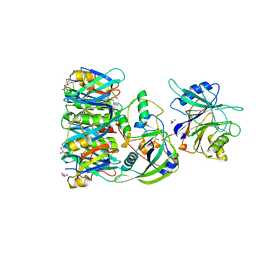 | | Structure of Typhoid Toxin | | Descriptor: | Cytolethal distending toxin subunit B homolog, GLYCEROL, Putative pertussis-like toxin subunit | | Authors: | Gao, X, Song, J, Galan, J. | | Deposit date: | 2013-04-16 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structure and function of the Salmonella Typhi chimaeric A(2)B(5) typhoid toxin.
Nature, 499, 2013
|
|
5YBH
 
 | |
6PWC
 
 | | A complex structure of arrestin-2 bound to neurotensin receptor 1 | | Descriptor: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | Authors: | Yin, W, Li, Z, Jin, M, Yin, Y.-L, de Waal, P.W, Pal, K, Gao, X, He, Y, Gao, J, Wang, X, Zhang, Y, Zhou, H, Melcher, K, Jiang, Y, Cong, Y, Zhou, X.E, Yu, X, Xu, H.E. | | Deposit date: | 2019-07-22 | | Release date: | 2019-12-04 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | A complex structure of arrestin-2 bound to a G protein-coupled receptor.
Cell Res., 29, 2019
|
|
3U30
 
 | | Crystal structure of a linear-specific Ubiquitin fab bound to linear ubiquitin | | Descriptor: | Heavy chain Fab, Light chain Fab, linear di-ubiquitin | | Authors: | Matsumoto, M.L, Dong, K.C, Yu, C, Phu, L, Gao, X, Hannoush, R.N, Hymowitz, S.G, Kirkpatrick, D.S, Dixit, V.M, Kelley, R.F. | | Deposit date: | 2011-10-04 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.428 Å) | | Cite: | Engineering and structural characterization of a linear polyubiquitin-specific antibody.
J.Mol.Biol., 418, 2012
|
|
8HM1
 
 | |
8HM2
 
 | |
8HM4
 
 | | Crystal structure of PPIase | | Descriptor: | Peptidylprolyl isomerase | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
8HM3
 
 | | Complex of PPIase-BfUbb | | Descriptor: | GLYCEROL, MAGNESIUM ION, Peptidylprolyl isomerase, ... | | Authors: | Xu, J.H, Chen, Z, Gao, X. | | Deposit date: | 2022-12-02 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Bacteroides fragilis ubiquitin homologue drives intraspecies bacterial competition in the gut microbiome.
Nat Microbiol, 9, 2024
|
|
1NOQ
 
 | | e-motif structure | | Descriptor: | 5'-D(*CP*CP*GP*CP*CP*G)-3' | | Authors: | Zheng, M, Huang, X, Smith, G.K, Yang, X, Gao, X. | | Deposit date: | 2003-01-16 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Genetically unstable CXG repeats are structurally dynamic and have a high propensity for folding.
An NMR and UV spectroscopic study.
J.Mol.Biol., 264, 1996
|
|
8ZC4
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC0
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC6
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.85 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC3
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC5
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC1
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, focused refinement of RBD region | | Descriptor: | Heavy chain of D1F6 Fab, Light chain of D1F6 Fab, Spike protein S1 | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZC2
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with D1F6 Fab, head-to-head aggregate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (7.82 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZBY
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (x2-4P) in complex with 3 D1F6 Fabs (0 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8ZBZ
 
 | | SARS-CoV-2 Omicron BA.2 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.71 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
1NP5
 
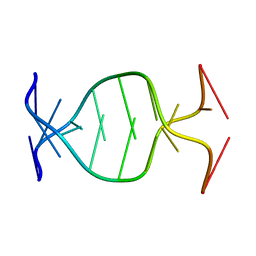 | | (GAC)3 parallel duplex | | Descriptor: | 5'-D(*GP*AP*CP*GP*AP*CP*GP*AP*C)-3' | | Authors: | Zheng, M, Han, X, Gao, X. | | Deposit date: | 2003-01-17 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Strand polarity of trinucleotide repeat sequences:
NMR studies of parallel/anti-pararell duplex,{d(GAC)3}2
To be Published
|
|
5XET
 
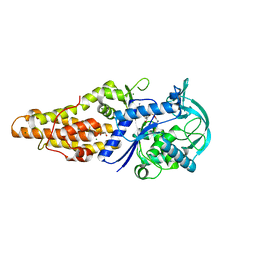 | | Crystal structure of Mycobacterium tuberculosis methionyl-tRNA synthetase bound by methionyl-adenylate (Met-AMP) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methionine--tRNA ligase, ... | | Authors: | Wang, W, Qin, B, Wojdyla, J.A, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2017-04-06 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural characterization of free-state and product-stateMycobacterium tuberculosismethionyl-tRNA synthetase reveals an induced-fit ligand-recognition mechanism.
IUCrJ, 5, 2018
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
1Q3H
 
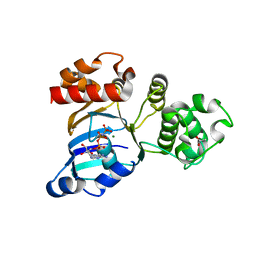 | | mouse CFTR NBD1 with AMP.PNP | | Descriptor: | ACETIC ACID, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, ... | | Authors: | Lewis, H.A, Buchanan, S.G, Burley, S.K, Conners, K, Dickey, M, Dorwart, M, Fowler, R, Gao, X, Guggino, W.B, Hendrickson, W.A. | | Deposit date: | 2003-07-29 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator.
Embo J., 23, 2004
|
|
