7YSW
 
 | | Cryo-EM Structure of FGF23-FGFR4-aKlotho-HS Quaternary Complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, COPPER (II) ION, Fibroblast growth factor 23, ... | | Authors: | Mohammadi, M, Chen, L. | | Deposit date: | 2022-08-13 | | Release date: | 2023-06-14 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for FGF hormone signalling.
Nature, 618, 2023
|
|
7YSU
 
 | | Cryo-EM Structure of FGF23-FGFR3c-aKlotho-HS Quaternary Complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, COPPER (II) ION, Fibroblast growth factor 23, ... | | Authors: | Mohammadi, M, Chen, L. | | Deposit date: | 2022-08-13 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for FGF hormone signalling.
Nature, 618, 2023
|
|
7YSH
 
 | | Cryo-EM Structure of FGF23-FGFR1c-aKlotho-HS Quaternary Complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, COPPER (II) ION, Fibroblast growth factor 23, ... | | Authors: | Mohammadi, M, Chen, L. | | Deposit date: | 2022-08-12 | | Release date: | 2023-04-19 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis for FGF hormone signalling.
Nature, 618, 2023
|
|
9B4U
 
 | | Crystal structure of p110alpha-RBD covalently bound to a breaker compound BBO-10203 via Cys242 | | Descriptor: | 1-[(4R,8R)-2-[(4M,7P)-7-[2,4-difluoro-6-(2-methoxyethoxy)phenyl]-4-(1-methyl-1H-indazol-5-yl)thieno[3,2-c]pyridin-6-yl]-4-methyl-6,7-dihydropyrazolo[1,5-a]pyrazin-5(4H)-yl]propan-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Czyzyk, D.J, Simanshu, D.K. | | Deposit date: | 2024-03-21 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | BBO-10203 inhibits tumor growth without inducing hyperglycemia by blocking RAS-PI3K alpha interaction.
Science, 2025
|
|
7YMI
 
 | | PSII-Pcb Dimer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a unique PSII-Pcb tetrameric megacomplex in a chlorophyll d -containing cyanobacterium.
Sci Adv, 10, 2024
|
|
8XFF
 
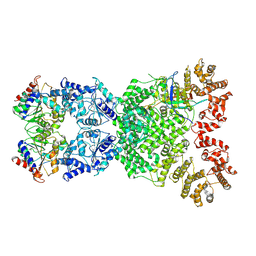 | | Cryo-EM structure of defence-associatedsirtuin 2 (DSR2) H171A protein in complex with SPR phage tail tube protein | | Descriptor: | DSR2(H171A), SPR tail tube protein | | Authors: | Li, Y, Zhang, H, Zheng, Q, Wu, Y, Li, S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insights into activation mechanisms on NADase of the bacterial DSR2 anti-phage defense system.
Sci Adv, 10, 2024
|
|
8XFE
 
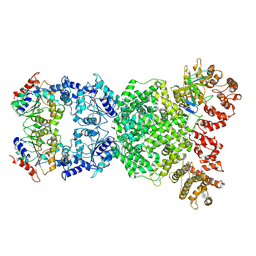 | | Cryo-EM structure of defence-associated sirtuin 2 (DSR2) H171A protein in complex with DSR anti-defence 1(DSAD1) | | Descriptor: | DSAD1, DSR2(H171A) | | Authors: | Li, Y, Zhang, H, Zheng, Q, Wu, Y, Li, S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural insights into activation mechanisms on NADase of the bacterial DSR2 anti-phage defense system.
Sci Adv, 10, 2024
|
|
4KMH
 
 | |
4KM9
 
 | |
4KMD
 
 | | Crystal structure of Sufud60-Gli1p | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, Sufu, ... | | Authors: | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | Deposit date: | 2013-05-08 | | Release date: | 2013-11-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
4KM8
 
 | | Crystal structure of Sufud60 | | Descriptor: | Suppressor of fused homolog | | Authors: | Zhang, Y, Qi, X, Zhang, Z, Wu, G. | | Deposit date: | 2013-05-08 | | Release date: | 2013-11-20 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci.
Nat Commun, 4, 2013
|
|
6IDA
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTQ ( A138S, P221T, L226Q) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6ID8
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTL ( A138S, P221T) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6ICX
 
 | |
6ID9
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SGTL ( A138S, V186G, P221T) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6ICW
 
 | |
6ID5
 
 | |
6ICY
 
 | | Crystal structure of H7 hemagglutinin mutant H7-AGTL ( V186G, P221T) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-07 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6IDB
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTQ ( A138S, P221T, L226Q) with 6'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6IDZ
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SVTQ ( A138S, P221T, L226Q) with 3'SLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain, ... | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-12 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.707 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
7XCP
 
 | | Cryo-EM structure of Omicron RBD complexed with ACE2 and 304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 304 Fab, ... | | Authors: | Zhao, Z, Qi, J, Gao, F.G. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape
Nat Commun, 13, 2022
|
|
6ID2
 
 | |
6ID3
 
 | | Crystal structure of H7 hemagglutinin mutant H7-SGPL ( A138S, V186G) from the influenza virus A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, Hemagglutinin HA2 chain | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-08 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
6IDD
 
 | | Crystal structure of H7 hemagglutinin mutant SH1-AVPL ( S138A, G186V, T221P, Q226L) from the influenza virus A/Shanghai/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Gao, G.F, Xu, Y, Qi, J.X. | | Deposit date: | 2018-09-09 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Avian-to-Human Receptor-Binding Adaptation of Avian H7N9 Influenza Virus Hemagglutinin.
Cell Rep, 29, 2019
|
|
8IEM
 
 | | Cryo-EM structure of ATP13A2 in the E2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Polyamine-transporting ATPase 13A2, ... | | Authors: | Liu, Z.M, Mu, J.Q, Xue, C.Y. | | Deposit date: | 2023-02-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Conformational cycle of human polyamine transporter ATP13A2.
Nat Commun, 14, 2023
|
|
