7ROA
 
 | | Crystal structure of EntV136 from Enterococcus faecalis | | Descriptor: | EntV | | Authors: | Stogios, P.J, Evdokimova, E, Kim, Y, Garsin, D, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-07-30 | | Release date: | 2022-10-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and functional analysis of EntV reveals a 12 amino acid fragment protective against fungal infections.
Nat Commun, 13, 2022
|
|
7S2M
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, Sul3 | | Authors: | Stogios, P.J, Skarina, T, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2I
 
 | | Crystal structure of sulfonamide resistance enzyme Sul1 in complex with 6-hydroxymethylpterin | | Descriptor: | 6-HYDROXYMETHYLPTERIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Kim, Y, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2J
 
 | | Crystal structure of sulfonamide resistance enzyme Sul2 apoenzyme | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2K
 
 | | Crystal structure of sulfonamide resistance enzyme Sul2 in complex with 7,8-dihydropteroate, magnesium, and pyrophosphate | | Descriptor: | 4-AMINOBENZOIC ACID, 7,8-DIHYDROPTEROATE, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2L
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 apoenzyme | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-09-03 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
9EDY
 
 | | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 2b: 6-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine | | Descriptor: | 6-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine, CHLORIDE ION, non-specific serine/threonine protein kinase | | Authors: | Stogios, P.J, Whitesell, L, Cowen, L.E, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-11-18 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 2b: 6-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine
To Be Published
|
|
9EDX
 
 | | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 2a: 2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine-6-carbonitrile | | Descriptor: | 2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine-6-carbonitrile, CHLORIDE ION, non-specific serine/threonine protein kinase | | Authors: | Stogios, P.J, Whitesell, L, Cowen, L.E, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-11-18 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 2a: 2-(4-fluorophenyl)-3-(pyridin-4-yl)pyrazolo[1,5-a]pyridine-6-carbonitrile
To Be Published
|
|
9EDW
 
 | | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 1f: 7-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine | | Descriptor: | 7-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine, CHLORIDE ION, Non-specific serine/threonine protein kinase | | Authors: | Stogios, P.J, Whitesell, L, Cowen, L.E, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-11-18 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 1f: 7-fluoro-2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine
To Be Published
|
|
8SQN
 
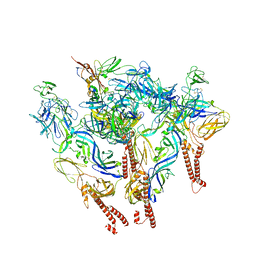 | | CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DuD1MoD2 chimeric MXRA8, E1 envelope glycoprotein, ... | | Authors: | Zimmerman, M.I, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-05-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Vertebrate-class-specific binding modes of the alphavirus receptor MXRA8.
Cell, 186, 2023
|
|
8F4Y
 
 | | Crystal Structure of SARS-CoV-2 2'-O-Methyltransferase in Complex with Compound 5a covalently bound to nsp16 and nsp10 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, 4-[2-(2,4-dichlorophenyl)ethyl]-6-(trifluoromethyl)pyrimidin-2-ol, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8F4S
 
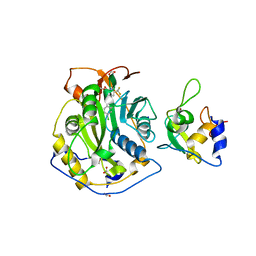 | | Crystal Structure of the SARS-CoV-2 2'-O-Methyltransferase with Compound 5a bound to the Cryptic Pocket of nsp16 | | Descriptor: | 2'-O-methyltransferase, 4-[(E)-2-(2,4-dichlorophenyl)ethenyl]-6-(trifluoromethyl)pyrimidin-2-ol, FORMIC ACID, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-11 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of a Druggable, Cryptic Pocket in SARS-CoV-2 nsp16 Using Allosteric Inhibitors.
Acs Infect Dis., 9, 2023
|
|
8F09
 
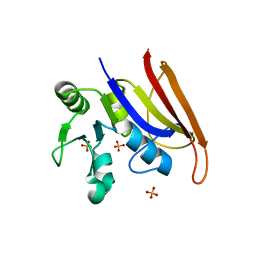 | | Crystal structure of a trimethoprim-resistant dihydrofolate reductase (DHFR) enzyme from an uncultured soil bacterium | | Descriptor: | Dihydrofolate reductase, SULFATE ION | | Authors: | Stogios, P.J, Evdokimova, D, Borek, D, Di Leo, R, Semper, C, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a trimethoprim-resistant dihydrofolate reductase (DHFR) enzyme from an uncultured soil bacterium
To Be Published
|
|
9AVI
 
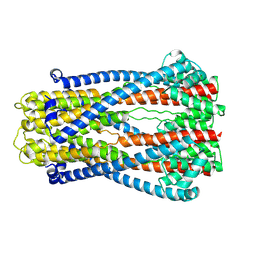 | |
8T28
 
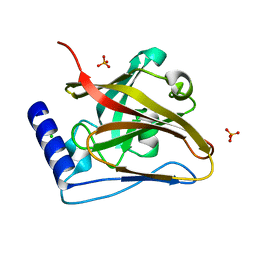 | | The crystal structure of SrtC2 sortase from Actinomyces oris | | Descriptor: | CHLORIDE ION, Class C sortase, PHOSPHATE ION | | Authors: | Osipiuk, J, Chang, C, Ton-That, H.L, Ton-That, H, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-06-05 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for dual functions in pilus assembly modulated by the lid of a pilus-specific sortase.
J.Biol.Chem., 300, 2024
|
|
9EDV
 
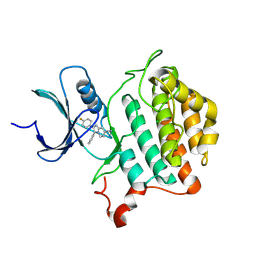 | | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 1e: 2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine-7-carbonitrile | | Descriptor: | 2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine-7-carbonitrile, CHLORIDE ION, non-specific serine/threonine protein kinase | | Authors: | Stogios, P.J, Whitesell, L, Cowen, L.E, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-11-18 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of Yck2 from Candida albicans in complex with inhibitor 1e: 2-(4-fluorophenyl)-3-(pyridin-4-yl)imidazo[1,2-a]pyridine-7-carbonitrile
To Be Published
|
|
8UFB
 
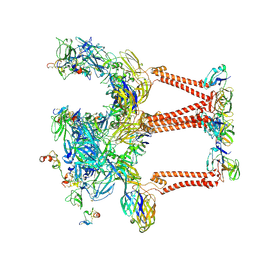 | |
8UFC
 
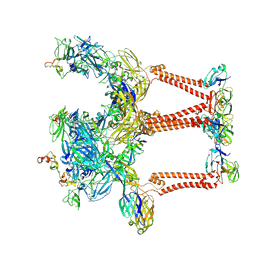 | |
8UFA
 
 | |
9BHC
 
 | |
9BHE
 
 | |
9CLY
 
 | | Crystal structure of the 3-ketoacyl-(acyl-carrier-protein) reductase, CylG, from Streptococcus agalactiae 2603V/R | | Descriptor: | CylG protein, IMIDAZOLE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Maltseva, N, Kim, Y, Endres, M, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-07-12 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the 3-ketoacyl-(acyl-carrier-protein) reductase, CylG, from Streptococcus agalactiae 2603V/R
To be published
|
|
9DDY
 
 | | The Crystal Structure of Geranyltranstransferase from Streptococcus pneumoniae TIGR4 | | Descriptor: | GLYCEROL, Geranyltranstransferase | | Authors: | Kim, Y, Tan, A, Endres, M, Tan, K, Joachimik, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-08-28 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | The Crystal Structure of Geranyltranstransferase from Streptococcus pneumoniae TIGR4
To Be Published
|
|
9D91
 
 | | Crystal structure of L-asparaginase from Streptococcus pneumoniae TIGR4 | | Descriptor: | 1,2-ETHANEDIOL, Asparaginase, SULFATE ION | | Authors: | Gade, P, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-08-20 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of L-asparaginase from
Streptococcus pneumoniae TIGR4
To Be Published
|
|
8UW6
 
 | | Acetylornithine deacetylase from Escherichia coli, di-zinc form. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetylornithine deacetylase, ... | | Authors: | Osipiuk, J, Endres, M, Kelley, E, Becker, D.P, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-11-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N alpha-acetyl-L-ornithine deacetylase from Escherichia coli and a ninhydrin-based assay to enable inhibitor identification.
Front Chem, 12, 2024
|
|
