8S5F
 
 | | Crystal structure of the HExxH domain of ChlBHExxH a novel alpha-ketoglutarate dependent oxygenase | | Descriptor: | ChlH from Chlorogloeopsis sp., PHOSPHATE ION | | Authors: | de la Mora, E, Amara, P, Usclat, A, Morishita, Y, Morinaka, B, Nicolet, Y. | | Deposit date: | 2024-02-23 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Fused radical SAM and alpha KG-HExxH domain proteins contain a distinct structural fold and catalyse cyclophane formation and beta-hydroxylation.
Nat.Chem., 16, 2024
|
|
5HLG
 
 | | Structure of reduced AbfR bound to DNA | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*TP*CP*AP*AP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*TP*TP*GP*AP*GP*T)-3'), MarR family transcriptional regulator | | Authors: | Liu, G, Liu, X, Gan, J, Yang, C.G. | | Deposit date: | 2016-01-15 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the Redox-Sensing Mechanism of MarR-Type Regulator AbfR.
J. Am. Chem. Soc., 139, 2017
|
|
8TS0
 
 | |
5HLI
 
 | | Structure of Disulfide formed AbfR | | Descriptor: | CHLORIDE ION, COPPER (II) ION, MarR family transcriptional regulator | | Authors: | Liu, G, Liu, X, Gan, J, Yang, C.-G. | | Deposit date: | 2016-01-15 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into the Redox-Sensing Mechanism of MarR-Type Regulator AbfR.
J. Am. Chem. Soc., 139, 2017
|
|
5HLH
 
 | | Crystal structure of the overoxidized AbfR bound to DNA | | Descriptor: | DNA (5'-D(*TP*AP*AP*CP*TP*CP*AP*AP*TP*CP*GP*CP*GP*CP*GP*CP*GP*AP*TP*TP*GP*AP*GP*T)-3'), MarR family transcriptional regulator | | Authors: | Liu, G, Liu, X, Gan, J, Yang, C.-G. | | Deposit date: | 2016-01-15 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into the Redox-Sensing Mechanism of MarR-Type Regulator AbfR.
J. Am. Chem. Soc., 139, 2017
|
|
6LW2
 
 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | Descriptor: | 7-chloranyl-4-[(3-methoxyphenyl)amino]-N-(4-methoxyphenyl)sulfonyl-1-methyl-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2020-02-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of N -Arylsulfonyl-Indole-2-Carboxamide Derivatives as Potent, Selective, and Orally Bioavailable Fructose-1,6-Bisphosphatase Inhibitors-Design, Synthesis, In Vivo Glucose Lowering Effects, and X-ray Crystal Complex Analysis.
J.Med.Chem., 63, 2020
|
|
6LRU
 
 | | Marsupenaeus japonicus ferritin mutant (T158H) | | Descriptor: | FE (III) ION, Ferritin, IMIDAZOLE, ... | | Authors: | Zhao, G, Tan, X, Zhang, T. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Converting histidine-induced 3D protein arrays in crystals into their 3D analogues in solution by metal coordination cross-linking.
Commun Chem, 2020
|
|
5GOU
 
 | | Structure of a 16-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | Descriptor: | Ferritin heavy chain | | Authors: | Zhang, S, Zang, J, Wang, W, Wang, H, Zhao, G. | | Deposit date: | 2016-07-29 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Conversion of the Native 24-mer Ferritin Nanocage into Its Non-Native 16-mer Analogue by Insertion of Extra Amino Acid Residues.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
1C9B
 
 | |
8W88
 
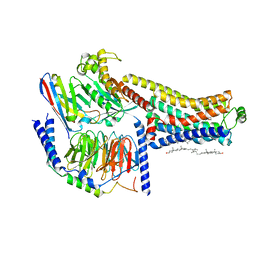 | | Cryo-EM structure of the SEP363856-bound TAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W8B
 
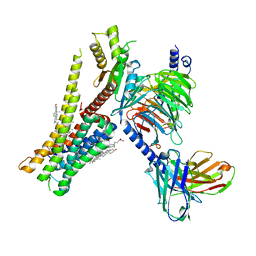 | | Cryo-EM structure of SEP-363856 bounded serotonin 1A (5-HT1A) receptor-Gi protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Antibody fragment scFv16, CHOLESTEROL, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W89
 
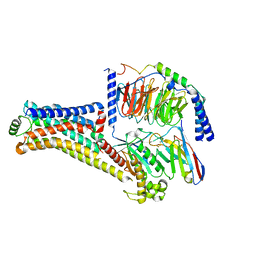 | | Cryo-EM structure of the PEA-bound TAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W87
 
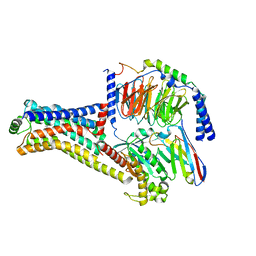 | | Cryo-EM structure of the METH-TAAR1 complex | | Descriptor: | (2S)-N-methyl-1-phenylpropan-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8W8A
 
 | | Cryo-EM structure of the RO5256390-TAAR1 complex | | Descriptor: | (4S)-4-[(2S)-2-phenylbutyl]-1,3-oxazolidin-2-imine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, H, Zheng, Y, Wang, Y, Wang, Y, He, X, Xu, P, Huang, S, Yuan, Q, Zhang, X, Wang, S, Xu, H.E, Xu, F. | | Deposit date: | 2023-09-01 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Recognition of methamphetamine and other amines by trace amine receptor TAAR1.
Nature, 624, 2023
|
|
6NW2
 
 | | Structure of human RIPK1 kinase domain in complex with compound 11 | | Descriptor: | (5R)-5-methyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-4,5,6,7-tetrahydro-2H-indazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and selective inhibitors of receptor-interacting protein kinase 1 that lack an aromatic back pocket group.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
5HEK
 
 | | crystal structure of M1.HpyAVI | | Descriptor: | Adenine specific DNA methyltransferase (DpnA) | | Authors: | Ma, B, Zhang, H, Liu, W. | | Deposit date: | 2016-01-06 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical and structural characterization of a DNA N6-adenine methyltransferase from Helicobacter pylori
Oncotarget, 7, 2016
|
|
6LRV
 
 | |
5HFJ
 
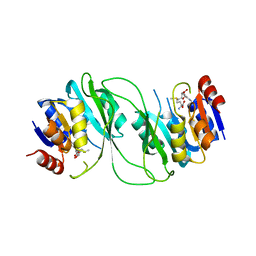 | | crystal structure of M1.HpyAVI-SAM complex | | Descriptor: | Adenine specific DNA methyltransferase (DpnA), S-ADENOSYLMETHIONINE | | Authors: | Ma, B, Liu, W, Zhang, H. | | Deposit date: | 2016-01-07 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical and structural characterization of a DNA N6-adenine methyltransferase from Helicobacter pylori
Oncotarget, 7, 2016
|
|
7XDX
 
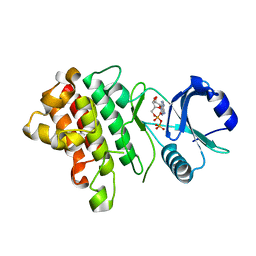 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7XDV
 
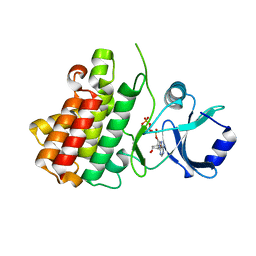 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7XDY
 
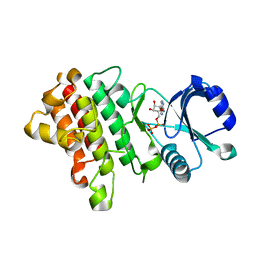 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7XDW
 
 | | Crystal structure of a receptor like kinase from Arabidopsis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Receptor-like protein kinase FERONIA | | Authors: | Kong, Y.Q, Ming, Z.H. | | Deposit date: | 2022-03-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.926 Å) | | Cite: | Structural and biochemical basis of Arabidopsis FERONIA receptor kinase-mediated early signaling initiation.
Plant Commun., 4, 2023
|
|
7X1H
 
 | |
7X1G
 
 | |
7X1J
 
 | | Cryo-EM structure of human BTR1 in the outward-facing state in the presence of NH4Cl. | | Descriptor: | Isoform 1 of Solute carrier family 4 member 11, [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate | | Authors: | Yin, Y, Lu, Y, Zuo, P. | | Deposit date: | 2022-02-24 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural insights into the conformational changes of BTR1/SLC4A11 in complex with PIP 2.
Nat Commun, 14, 2023
|
|
