7F59
 
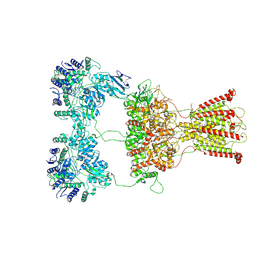 | | DNQX-bound GluK2-1xNeto2 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F56
 
 | | DNQX-bound GluK2-1xNeto2 complex, with asymmetric LBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F57
 
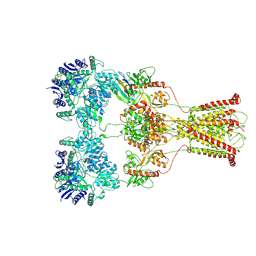 | | Kainate-bound GluK2-1xNeto2 complex, at the desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F5B
 
 | | LBD-TMD focused reconstruction of DNQX-bound GluK2-1xNeto2 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7F5A
 
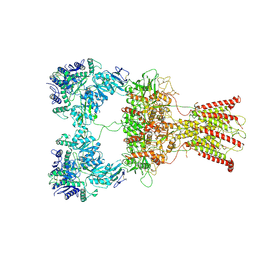 | | DNQX-bound GluK2-2xNeto2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7BEO
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-253H55L and COVOX-75 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEP
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-384 and S309 Fabs | | Descriptor: | CHLORIDE ION, COVOX-384 heavy chain, COVOX-384 light chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEI
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-150 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-150 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEJ
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-158 Fab (crystal form 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEL
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-88 and COVOX-45 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COVOX-45 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7DSV
 
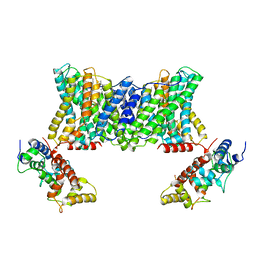 | | Structure of a human NHE1-CHP1 complex under pH 6.5 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Calcineurin B homologous protein 1, Sodium/hydrogen exchanger 1 | | Authors: | Dong, Y, Gao, Y, Li, B, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and mechanism of the human NHE1-CHP1 complex.
Nat Commun, 12, 2021
|
|
7DSW
 
 | | Structure of a human NHE1-CHP1 complex under pH 7.5 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Sodium/hydrogen exchanger 1 | | Authors: | Dong, Y, Gao, Y, Li, B, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and mechanism of the human NHE1-CHP1 complex.
Nat Commun, 12, 2021
|
|
7DSX
 
 | | Structure of a human NHE1-CHP1 complex under pH 7.5, bound by cariporide | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Calcineurin B homologous protein 1, N-[bis(azanyl)methylidene]-3-methylsulfonyl-4-propan-2-yl-benzamide, ... | | Authors: | Dong, Y, Gao, Y, Li, B, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and mechanism of the human NHE1-CHP1 complex.
Nat Commun, 12, 2021
|
|
7EJ2
 
 | | human voltage-gated potassium channel KV1.3 H451N mutant | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 3, Voltage-gated potassium channel subunit beta-2 | | Authors: | Liu, S, Zhao, Y, Tian, C. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of wild-type and H451N mutant human lymphocyte potassium channel K V 1.3.
Cell Discov, 7, 2021
|
|
7EJ1
 
 | | human voltage-gated potassium channel KV1.3 | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 3, Voltage-gated potassium channel subunit beta-2 | | Authors: | Liu, S, Zhao, Y, Tian, C. | | Deposit date: | 2021-04-01 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of wild-type and H451N mutant human lymphocyte potassium channel K V 1.3.
Cell Discov, 7, 2021
|
|
7BEM
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-269 scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-269 Vh domain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEH
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-316 Fab | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-316 heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEN
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in a ternary complex with COVOX-253 and COVOX-75 Fabs | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-24 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7BEK
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-158 Fab (crystal form 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhou, D, Zhao, Y, Ren, J, Stuart, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7F3U
 
 | | Cryo-EM structure of human TMEM120A in the CoASH-free state | | Descriptor: | Transmembrane protein 120A | | Authors: | Rong, Y, Gao, Y.W, Song, D.F, Zhao, Y, Liu, Z.F. | | Deposit date: | 2021-06-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | TMEM120A contains a specific coenzyme A-binding site and might not mediate poking- or stretch-induced channel activities in cells.
Elife, 10, 2021
|
|
7CUT
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with Z-VAD-FMK | | Descriptor: | 3C protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lu, M, Yang, H.T, Wang, Z.Y, Zhao, Y, Xing, Y.F. | | Deposit date: | 2020-08-24 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Identification of proteasome and caspase inhibitors targeting SARS-CoV-2 M pro .
Signal Transduct Target Ther, 6, 2021
|
|
7CUU
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with MG132 | | Descriptor: | 3C protein, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lu, M, Yang, H.T, Wang, Z.Y, Zhao, Y, Xing, Y.F. | | Deposit date: | 2020-08-24 | | Release date: | 2021-06-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification of proteasome and caspase inhibitors targeting SARS-CoV-2 M pro .
Signal Transduct Target Ther, 6, 2021
|
|
7WU7
 
 | | Prefoldin-tubulin-TRiC complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Prefoldin subunit 1, Prefoldin subunit 2, ... | | Authors: | Gestaut, D, Zhao, Y, Park, J, Ma, B, Leitner, A, Collier, M, Pintilie, G, Roh, S.-H, Chiu, W, Frydman, J. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7WF3
 
 | | Composite map of human Kv1.3 channel in apo state with beta subunits | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, ... | | Authors: | Tyagi, A, Ahmed, T, Jian, S, Bajaj, S, Ong, S.T, Goay, S.S.M, Zhao, Y, Vorobyov, I, Tian, C, Chandy, K.G, Bhushan, S. | | Deposit date: | 2021-12-25 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rearrangement of a unique Kv1.3 selectivity filter conformation upon binding of a drug.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WF4
 
 | | Composite map of human Kv1.3 channel in dalazatide-bound state with beta subunits | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, ... | | Authors: | Tyagi, A, Ahmed, T, Jian, S, Bajaj, S, Ong, S.T, Goay, S.S.M, Zhao, Y, Vorobyov, I, Tian, C, Chandy, K.G, Bhushan, S. | | Deposit date: | 2021-12-25 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rearrangement of a unique Kv1.3 selectivity filter conformation upon binding of a drug.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
