5C31
 
 | |
5C35
 
 | |
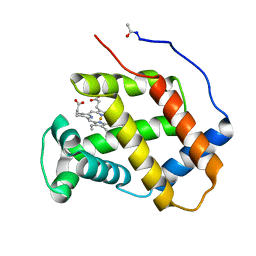
5CE4
 
 | | High Resolution X-Ray and Neutron diffraction structure of H-FABP | | Descriptor: | Fatty acid-binding protein, heart, OLEIC ACID | | Authors: | Podjarny, A.D, Howard, E.I, Blakeley, M.P, Guillot, B. | | Deposit date: | 2015-07-06 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (0.98 Å), X-RAY DIFFRACTION | | Cite: | High-resolution neutron and X-ray diffraction room-temperature studies of an H-FABP-oleic acid complex: study of the internal water cluster and ligand binding by a transferred multipolar electron-density distribution.
Iucrj, 3, 2016
|
|
1HLB
 
 | |
1KTQ
 
 | | DNA POLYMERASE | | Descriptor: | DNA POLYMERASE I | | Authors: | Korolev, S, Waksman, G. | | Deposit date: | 1995-08-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the large fragment of Thermus aquaticus DNA polymerase I at 2.5-A resolution: structural basis for thermostability.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
3HDK
 
 | | Crystal structure of chemically synthesized [Aib51/51']HIV-1 protease | | Descriptor: | N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, [Aib51/51']HIV-1 protease | | Authors: | Torbeev, V.Y, Kent, S.B.H. | | Deposit date: | 2009-05-07 | | Release date: | 2010-04-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein conformational dynamics in the mechanism of HIV-1 protease catalysis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3IAW
 
 | |
3INT
 
 | | Structure of UDP-galactopyranose mutase bound to UDP-galactose (reduced) | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GALACTOSE-URIDINE-5'-DIPHOSPHATE, Probable UDP-galactopyranose mutase, ... | | Authors: | Gruber, T.D, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2009-08-12 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | X-ray crystallography reveals a reduced substrate complex of UDP-galactopyranose mutase poised for covalent catalysis by flavin .
Biochemistry, 48, 2009
|
|
3IMX
 
 | | Crystal Structure of human glucokinase in complex with a synthetic activator | | Descriptor: | (2R)-3-cyclopentyl-N-(5-methoxy[1,3]thiazolo[5,4-b]pyridin-2-yl)-2-{4-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}propanamide, Glucokinase, SODIUM ION, ... | | Authors: | Stams, T, Vash, B. | | Deposit date: | 2009-08-11 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of functionally liver selective glucokinase activators for the treatment of type 2 diabetes.
J.Med.Chem., 52, 2009
|
|
3GGX
 
 | | HIV Protease, pseudo-symmetric inhibitors | | Descriptor: | V-1 protease, methyl [(1S)-1-{[(1R,3S,4S)-4-{[(2S)-3,3-dimethyl-2-{3-[(6-methylpyridin-2-yl)methyl]-2-oxo-2,3-dihydro-1H-imidazol-1-yl}butanoyl]amino}-3-hydroxy-5-phenyl-1-(4-pyridin-2-ylbenzyl)pentyl]carbamoyl}-2,2-dimethylpropyl]carbamate | | Authors: | Stoll, V.S. | | Deposit date: | 2009-03-02 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus
protease inhibitors (A-792611 and A-790742) with potential for convenient
dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
3FRS
 
 | | Structure of human IST1(NTD) (residues 1-189)(p43212) | | Descriptor: | GLYCEROL, Uncharacterized protein KIAA0174 | | Authors: | Schubert, H.L, Hill, C.P, Bajorek, M, Sundquist, W.I. | | Deposit date: | 2009-01-08 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for ESCRT-III protein autoinhibition.
Nat.Struct.Mol.Biol., 16, 2009
|
|
8YZD
 
 | | Structure of JN.1 RBD protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,fusion protein | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZE
 
 | | The JN.1 spike protein (S) in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZC
 
 | | Structure of BA.2.86 spike protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,Expression Tag | | Authors: | Wang, Y.J, Zang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZB
 
 | | BA.2.86 RBD protein in complex with ACE2. | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,fusion protein | | Authors: | Wang, Y.J, Zhang, X, Sun, L. | | Deposit date: | 2024-04-06 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
3FRV
 
 | |
5ZRT
 
 | | Crystal structure of human C1ORF123 protein | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rahaman, S.N.A, Yusop, J.M, Mohamed-Hussein, Z.A, Wan Mohd, A, Ho, K.L, Teh, A.H, Waterman, J, Ng, C.L. | | Deposit date: | 2018-04-25 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of human C1ORF123.
Peerj, 6, 2018
|
|
8YWD
 
 | |
3INR
 
 | |
3GF4
 
 | | Structure of UDP-galactopyranose mutase bound to UDP-glucose | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, UDP-galactopyranose mutase, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | Authors: | Gruber, T.D, Borrok, M.J, Kiessling, L.L, Forest, K.T. | | Deposit date: | 2009-02-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Ligand binding and substrate discrimination by UDP-galactopyranose mutase.
J.Mol.Biol., 391, 2009
|
|
3HLO
 
 | |
6ABH
 
 | |
3GGA
 
 | | HIV Protease inhibitors with pseudo-symmetric cores | | Descriptor: | V-1 protease, methyl [(1S,4S,5S,7S,10S)-4-benzyl-1,10-di-tert-butyl-5-hydroxy-2,9,12-trioxo-7-(4-pyridin-2-ylbenzyl)-13-oxa-3,8,11-triazatetradec-1-yl]carbamate | | Authors: | Stoll, V.S. | | Deposit date: | 2009-02-27 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus
protease inhibitors (A-792611 and A-790742) with potential for convenient
dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
8Z2L
 
 | |
8Z2Q
 
 | |
