9JF4
 
 | | cryo-EM structure of Neuromedin B receptor in complex with PD168368 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-2-methyl-2-[(4-nitrophenyl)carbamoylamino]-~{N}-[(1-pyridin-2-ylcyclohexyl)methyl]propanamide, Neuromedin-B receptor,de novo design protein | | Authors: | Gao, K, Zhang, X, Liu, X. | | Deposit date: | 2024-09-03 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | cryo-EM structure of Neuromedin B receptor in complex with PD168368
To Be Published
|
|
7M0R
 
 | | Cryo-EM structure of the Sema3A/PlexinA4/Neuropilin 1 complex | | Descriptor: | CALCIUM ION, Neuropilin-1, Plexin-A4, ... | | Authors: | Lu, D, Shang, G, He, X, Bai, X, Zhang, X. | | Deposit date: | 2021-03-11 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Architecture of the Sema3A/PlexinA4/Neuropilin tripartite complex.
Nat Commun, 12, 2021
|
|
5ADY
 
 | | Cryo-EM structures of the 50S ribosome subunit bound with HflX | | Descriptor: | 23S RRNA, 50S RIBOSOMAL PROTEIN L1, 50S RIBOSOMAL PROTEIN L10, ... | | Authors: | Zhang, Y, Mandava, C.S, Cao, W, Li, X, Zhang, D, Li, N, Zhang, Y, Zhang, X, Qin, Y, Mi, K, Lei, J, Sanyal, S, Gao, N. | | Deposit date: | 2015-08-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Hflx is a Ribosome Splitting Factor Rescuing Stalled Ribosomes Under Stress Conditions
Nat.Struct.Mol.Biol., 22, 2015
|
|
5IJC
 
 | | The crystal structure of mouse TLR4/MD-2/neoseptin-3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJB
 
 | | The ligand-free structure of the mouse TLR4/MD-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJD
 
 | | The crystal structure of mouse TLR4/MD-2/lipid A complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1AWP
 
 | | RAT OUTER MITOCHONDRIAL MEMBRANE CYTOCHROME B5 | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, X, Zhang, X. | | Deposit date: | 1997-10-03 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The reduction potential of cytochrome b5 is modulated by its exposed heme edge.
Biochemistry, 37, 1998
|
|
1C78
 
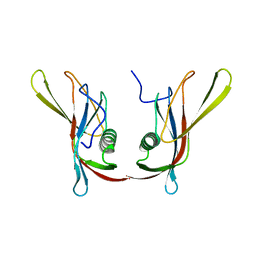 | | STAPHYLOKINASE (SAK) DIMER | | Descriptor: | STAPHYLOKINASE | | Authors: | Rao, Z, Jiang, F, Liu, Y, Zhang, X, Chen, Y, Bartlam, M, Song, H, Ding, Y. | | Deposit date: | 2000-02-01 | | Release date: | 2000-08-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a staphylokinase: variant a model for reduced antigenicity.
Eur.J.Biochem., 269, 2002
|
|
3ZN8
 
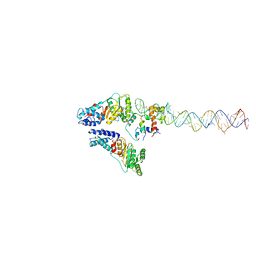 | | Structural Basis of Signal Sequence Surveillance and Selection by the SRP-SR Complex | | Descriptor: | 4.5 S RNA, DIPEPTIDYL AMINOPEPTIDASE B, MAGNESIUM ION, ... | | Authors: | von Loeffelholz, O, Knoops, K, Ariosa, A, Zhang, X, Karuppasamy, M, Huard, K, Schoehn, G, Berger, I, Shan, S.O, Schaffitzel, C. | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural Basis of Signal Sequence Surveillance and Selection by the Srp-Sr Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
6Z2W
 
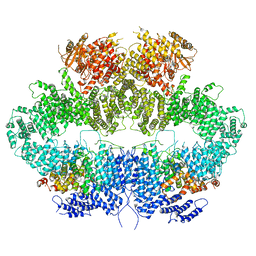 | | Mec1-Ddc2 (F2244L mutant) in complex with Mg AMP-PNP | | Descriptor: | DNA damage checkpoint protein LCD1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2020-05-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Mechanism of auto-inhibition and activation of Mec1 ATR checkpoint kinase.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6Z2X
 
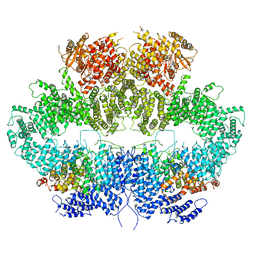 | | Mec1-Ddc2 (F2244L mutant) in complex with Mg AMP-PNP (State II) | | Descriptor: | DNA damage checkpoint protein LCD1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yates, L.A, Zhang, X. | | Deposit date: | 2020-05-18 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of auto-inhibition and activation of Mec1 ATR checkpoint kinase.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2UXU
 
 | | TtgR in complex with Naringenin | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGR, NARINGENIN | | Authors: | Alguel, Y, Meng, C, Teran, W, Krell, T, Ramos, J.L, Gallegos, M.-T, Zhang, X. | | Deposit date: | 2007-03-29 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Multidrug Binding Protein Ttgr in Complex with Antibiotics and Plant Antimicrobials.
J.Mol.Biol., 369, 2007
|
|
2UXO
 
 | | TtgR in complex with Tetracycline | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGR, TETRACYCLINE | | Authors: | Alguel, Y, Meng, C, Teran, W, Krell, T, Ramos, J.L, Gallegos, M.-T, Zhang, X. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Multidrug Binding Protein Ttgr in Complex with Antibiotics and Plant Antimicrobials.
J.Mol.Biol., 369, 2007
|
|
2UXI
 
 | | Phloretin in complex with TtgR | | Descriptor: | 3-(4-HYDROXYPHENYL)-1-(2,4,6-TRIHYDROXYPHENYL)PROPAN-1-ONE, HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGR | | Authors: | Alguel, Y, Meng, C, Teran, W, Krell, T, Ramos, J.L, Gallegos, M.-T, Zhang, X. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Multidrug Binding Protein Ttgr in Complex with Antibiotics and Plant Antimicrobials.
J.Mol.Biol., 369, 2007
|
|
2W5T
 
 | | Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. | | Descriptor: | (2R)-2,3-dihydroxypropyl phosphate, ACETATE ION, MANGANESE (II) ION, ... | | Authors: | Lu, D, Wormann, M.E, Zhang, X, Schneewind, O, Grundling, A, Freemont, P.S. | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Mechanism of Lipoteichoic Acid Synthesis by Staphylococcus Aureus Ltas.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6IRH
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class III | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6M3W
 
 | | Post-fusion structure of SARS-CoV spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Fan, X, Cao, D, Zhang, X. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-03 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein.
Nat Commun, 11, 2020
|
|
2WUI
 
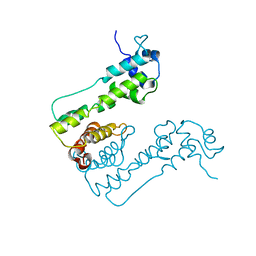 | | Crystal Structure of MexZ, a key repressor responsible for antibiotic resistance in Pseudomonas aeruginosa. | | Descriptor: | TRANSCRIPTIONAL REGULATOR | | Authors: | Alguel, Y, Lu, D, Quade, N, Zhang, X. | | Deposit date: | 2009-10-05 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Mexz, a Key Repressor Responsible for Antibiotic Resistance in Pseudomonas Aeruginosa.
J.Struct.Biol., 172, 2010
|
|
2XDN
 
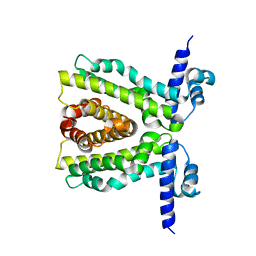 | |
2XRN
 
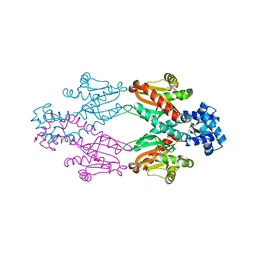 | | Crystal structure of TtgV | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGV | | Authors: | Lu, D, Fillet, S, Meng, C, Alguel, Y, Kloppsteck, P, Bergeron, J, Krell, T, Gallegos, M.-T, Ramos, J, Zhang, X. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Ttgv in Complex with its DNA Operator Reveals a General Model for Cooperative DNA Binding of Tetrameric Gene Regulators.
Genes Dev., 24, 2010
|
|
8DOL
 
 | | Mechanism of regulation of the Helicobacter pylori Cagbeta ATPase by CagZ | | Descriptor: | Cag pathogenicity island protein (Cag5), DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Wu, X, Zhao, Y, Yang, W, Sun, L, Ye, X, Jiang, M, Wang, Q, Wang, Q, Zhang, X, Wu, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of regulation of the Helicobacter pylori Cag beta ATPase by CagZ.
Nat Commun, 14, 2023
|
|
2W5Q
 
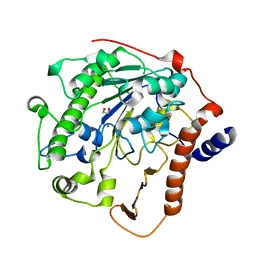 | | Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, PROCESSED GLYCEROL PHOSPHATE LIPOTEICHOIC ACID SYNTHASE | | Authors: | Lu, D, Wormann, M.E, Zhang, X, Scheewind, O, Grundling, A, Freemont, P.S. | | Deposit date: | 2008-12-11 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Based Mechanism of Lipoteichoic Acid Synthesis by Staphylococcus Aureus Ltas.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
