3FMQ
 
 | | Crystal structure of an Encephalitozoon cuniculi methionine aminopeptidase type 2 with angiogenesis inhibitor fumagillin bound | | Descriptor: | FE (III) ION, FUMAGILLIN, Methionine aminopeptidase 2, ... | | Authors: | Alvarado, J.J, Russell, M, Zhang, A, Adams, J, Toro, R, Burley, S.K, Weiss, L.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-22 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a microsporidian methionine aminopeptidase type 2 complexed with fumagillin and TNP-470.
Mol.Biochem.Parasitol., 168, 2009
|
|
3F5S
 
 | | CRYSTAL STRUCTURE OF putatitve short chain dehydrogenase from Shigella flexneri 2a str. 301 | | Descriptor: | dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CRYSTAL STRUCTURE OF putatitve short chain dehydrogenase from Shigella flexneri 2a str. 301
To be Published
|
|
3F6C
 
 | | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI | | Descriptor: | GLYCEROL, Positive transcription regulator evgA | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Wu, B, Bain, K, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-05 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CRYSTAL STRUCTURE OF N-TERMINAL DOMAIN OF POSITIVE TRANSCRIPTION REGULATOR evgA FROM ESCHERICHIA COLI
To be Published
|
|
3THU
 
 | | Crystal structure of an enolase from sphingomonas sp. ska58 (efi target efi-501683) with bound mg | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-08-19 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an enolase from sphingomonas sp. ska58 (efi target efi-501683) with bound mg
to be published
|
|
3FC0
 
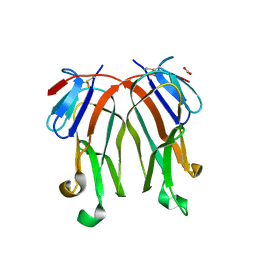 | | 1.8 A crystal structure of murine GITR ligand dimer expressed in Drosophila melanogaster S2 cells | | Descriptor: | ACETATE ION, GITR ligand | | Authors: | Chattopadhyay, K, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2008-11-20 | | Release date: | 2008-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 1.8 A structure of murine GITR ligand dimer expressed in Drosophila melanogaster S2 cells.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3FK4
 
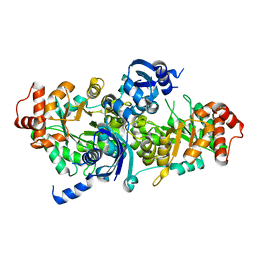 | | Crystal structure of RuBisCO-like protein from Bacillus Cereus ATCC 14579 | | Descriptor: | RuBisCO-like protein | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-15 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of RuBisCO-like protein from Bacillus Cereus ATCC 14579
To be Published
|
|
3FHL
 
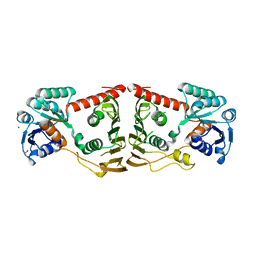 | | Crystal structure of a putative oxidoreductase from bacteroides fragilis nctc 9343 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative oxidoreductase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-09 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of a Putative Oxidoreductase from Bacteroides Fragilis Nctc 9343
To be Published
|
|
3TJI
 
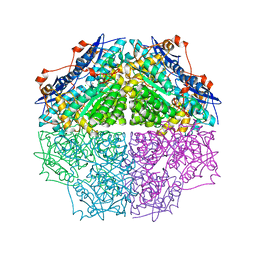 | | CRYSTAL STRUCTURE OF AN ENOLASE FROM ENTEROBACTER sp. 638 (EFI TARGET EFI-501662) with BOUND MG | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-08-24 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an enolase from enterobacter sp. 638 (efi target efi-501662) with boung mg
to be published
|
|
3VC7
 
 | | Crystal Structure of a putative oxidoreductase from Sinorhizobium meliloti 1021 | | Descriptor: | GLYCEROL, Putative oxidoreductase | | Authors: | Ghosh, A, Bhoshle, R, Toro, R, Gizzi, A, Hillerich, B, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | Crystal Structure of a putative oxidoreductase protein from Sinorhizobium meliloti 1021
To be Published
|
|
3VE7
 
 | | Crystal structure of orotidine 5'-monophosphate decarboxylase from Metallosphaera sedula complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, ACETIC ACID, Orotidine-5'-phosphate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-01-07 | | Release date: | 2013-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Crystal structure of orotidine 5'-monophosphate decarboxylase from Metallosphaera sedula complexed with inhibitor BMP
To be Published
|
|
3F5Q
 
 | | CRYSTAL STRUCTURE OF putative short chain dehydrogenase from Escherichia coli CFT073 | | Descriptor: | dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of an uncharacterized protein
to be published
|
|
3FDU
 
 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Acinetobacter baumannii | | Descriptor: | GLYCEROL, Putative enoyl-CoA hydratase/isomerase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Tang, B.K, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-26 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Acinetobacter baumannii
To be Published
|
|
3FEQ
 
 | | Crystal structure of uncharacterized protein eah89906 | | Descriptor: | PUTATIVE AMIDOHYDROLASE, ZINC ION | | Authors: | Patskovsky, Y, Bonanno, J, Romero, R, Freeman, J, Lau, C, Smith, D, Bain, K, Wasserman, S.R, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-30 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3FDK
 
 | | Crystal structure of hydrolase DR0930 with promiscuous catalytic activity | | Descriptor: | HYDROLASE DR0930, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, L.V, Xiang, D.F, Raushel, F.M, Almo, S.C. | | Deposit date: | 2008-11-25 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional annotation and three-dimensional structure of Dr0930 from Deinococcus radiodurans, a close relative of phosphotriesterase in the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3TJ4
 
 | | Crystal structure of an enolase from agrobacterium tumefaciens (efi target efi-502087) no mg | | Descriptor: | CHLORIDE ION, GLYCEROL, Mandelate racemase, ... | | Authors: | Vetting, M.W, Bouvier, J.T, Wasserman, S.R, Morisco, L.L, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of an enolase from agrobacterium tumefaciens (efi target efi-502087) no mg
to be published
|
|
3FON
 
 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide VNDIFEAI | | Descriptor: | Beta-2-microglobulin, MHC, Peptide | | Authors: | Malashkevich, V.N, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Brims, D.R, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | Deposit date: | 2008-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect.
Int.Immunol., 22, 2010
|
|
3TOY
 
 | | CRYSTAL STRUCTURE OF ENOLASE BRADO_4202 (TARGET EFI-501651) FROM Bradyrhizobium sp. ORS278 WITH CALCIUM AND ACETATE BOUND | | Descriptor: | ACETATE ION, CALCIUM ION, Mandelate racemase/muconate lactonizing enzyme family protein, ... | | Authors: | Patskovsky, Y, Kim, J, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammond, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-06 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF MANDELATE RACEMASE FROM Bradyrhizobium sp. ORS278
To be Published
|
|
3UN1
 
 | | Crystal structure of an oxidoreductase from Sinorhizobium meliloti 1021 | | Descriptor: | PHOSPHATE ION, Probable oxidoreductase | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-15 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of an oxidoreductase from Sinorhizobium meliloti 1021
To be Published
|
|
3FBG
 
 | | Crystal structure of a putative arginate lyase from Staphylococcus haemolyticus | | Descriptor: | MAGNESIUM ION, putative arginate lyase | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a putative arginate lyase from Staphylococcus haemolyticus
To be Published
|
|
3FNR
 
 | | CRYSTAL STRUCTURE OF PUTATIVE ARGINYL T-RNA SYNTHETASE FROM Campylobacter jejuni; | | Descriptor: | Arginyl-tRNA synthetase, GLYCEROL, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-26 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF A PUTATIVE ARGINYL T-RNA SYNTHETASE FROM Campylobacter jejuni
To be Published
|
|
3FOL
 
 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide VNDIFERI | | Descriptor: | 8 residue synthetic peptide, Beta-2-microglobulin, MHC | | Authors: | Brims, D.R, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Malashkevich, V.N, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | Deposit date: | 2008-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect
Int.Immunol., 22, 2010
|
|
3FCP
 
 | | Crystal structure of Muconate lactonizing enzyme from Klebsiella pneumoniae | | Descriptor: | L-Ala-D/L-Glu epimerase, a muconate lactonizing enzyme, MAGNESIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-22 | | Release date: | 2008-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Muconate lactonizing enzyme from Klebsiella pneumoniae
To be Published
|
|
3FJ4
 
 | | Crystal structure of muconate lactonizing enzyme from Pseudomonas Fluorescens complexed with muconolactone | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, [(2S)-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-14 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes
Biochemistry, 48, 2009
|
|
3UF0
 
 | | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Short-chain dehydrogenase/reductase SDR | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative NAD(P) dependent gluconate 5-dehydrogenase from Beutenbergia cavernae(EFI target EFI-502044) with bound NADP (low occupancy)
To be Published
|
|
3FI9
 
 | | Crystal structure of malate dehydrogenase from Porphyromonas gingivalis | | Descriptor: | Malate dehydrogenase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Miller, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-11 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of malate dehydrogenase from Porphyromonas gingivalis
To be Published
|
|
