8FNP
 
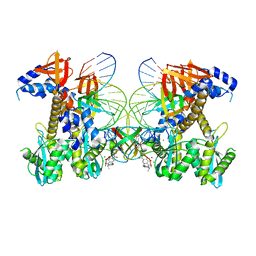 | | Structure of E138K/G140S/Q148H HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
9KMB
 
 | |
7Y01
 
 | |
7XF5
 
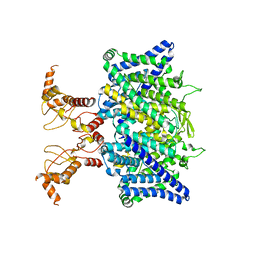 | | Full length human CLC-2 channel in apo state | | Descriptor: | Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-04-01 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42
Nat Commun, 14, 2023
|
|
7XJA
 
 | | TMD masked refine map of human ClC-2 | | Descriptor: | Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-04-15 | | Release date: | 2023-05-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42
Nat Commun, 14, 2023
|
|
5YJH
 
 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
7XRV
 
 | |
5YJG
 
 | | Structural insights into periostin functions | | Descriptor: | CALCIUM ION, CHLORIDE ION, CYSTEINE, ... | | Authors: | Liu, H, Liu, J, Xu, F. | | Deposit date: | 2017-10-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural characterizations of human periostin dimerization and cysteinylation.
FEBS Lett., 592, 2018
|
|
6ILG
 
 | |
4GQG
 
 | | Crystal structure of AKR1B10 complexed with NADP+ | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Hu, X. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
6IMC
 
 | | Crystal Structure of ALKBH1 in complex with Mn(II) and N-Oxalylglycine | | Descriptor: | MANGANESE (II) ION, N-OXALYLGLYCINE, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
6QFY
 
 | | CRYSTAL STRUCTURE OF PORCINE HEMAGGLUTINATING ENCEPHALOMYELITIS VIRUS SPIKE PROTEIN LECTIN DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Huizinga, E.G, Bakkers, M, Lang, Y. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6W9I
 
 | |
6W9H
 
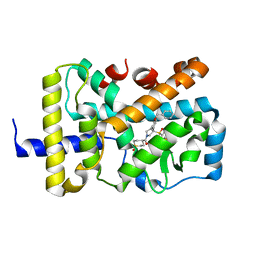 | | SUBSTITUTED BENZYLOXYTRICYCLIC COMPOUNDS AS RETINOIC ACID-RELATED ORPHAN RECEPTOR GAMMA T AGONISTS | | Descriptor: | 4-{(3R)-3-[4-(benzyloxy)phenyl]-3-[(4-fluorophenyl)sulfonyl]pyrrolidine-1-carbonyl}-1lambda~6~-thiane-1,1-dione, Nuclear receptor ROR-gamma, Steroid receptor coactivator 1 fusion | | Authors: | Sack, J.S. | | Deposit date: | 2020-03-23 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Substituted benzyloxytricyclic compounds as retinoic acid-related orphan receptor gamma t (ROR gamma t) agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6A0Z
 
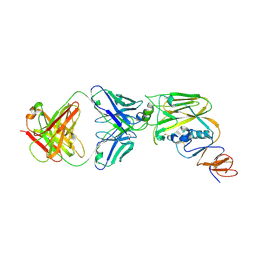 | | Crystal structure of broadly neutralizing antibody 13D4 bound to H5N1 influenza hemagglutinin, HA head region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 13D4, Fab Heavy Chain, ... | | Authors: | Li, S, Li, T. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | Structural Basis for the Broad, Antibody-Mediated Neutralization of H5N1 Influenza Virus.
J. Virol., 92, 2018
|
|
6VL8
 
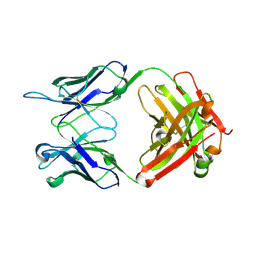 | | Anti-PEG antibody 6-3 Fab fragment in complex with PEG | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 6-3 Fab heavy chain, ... | | Authors: | Nicely, N.I, Huckaby, J.T, Lai, S.K, Jacobs, T.M. | | Deposit date: | 2020-01-23 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of an anti-PEG antibody reveals an open ring that captures highly flexible PEG polymers
Commun Chem, 3, 2020
|
|
6VL9
 
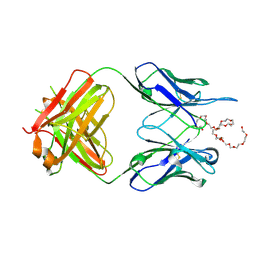 | | Anti-PEG antibody 6-3 Fab fragment in complex with PEG | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45,48,51,54,57-nonadecaoxanonapentacontane-1,59-diol, 6-3 Fab heavy chain, 6-3 Fab light chain | | Authors: | Nicely, N.I, Huckaby, J.T, Lai, S.K, Jacobs, T.M. | | Deposit date: | 2020-01-23 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure of an anti-PEG antibody reveals an open ring that captures highly flexible PEG polymers
Commun Chem, 3, 2020
|
|
9I4H
 
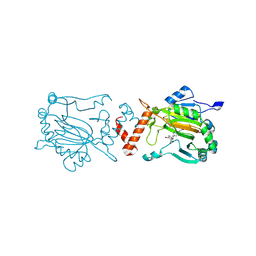 | | Factor Inhibiting HIF (FIH) in complex with manganese and 3-Hydroxy-5-(3-(4-(hydroxymethyl)-3-nitrophenyl)isoxazol-5-yl)picolinoyl)glycine | | Descriptor: | 2-[[5-[3-[4-(hydroxymethyl)-3-nitro-phenyl]-1,2-oxazol-5-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Hypoxia-inducible factor 1-alpha inhibitor, MANGANESE (II) ION | | Authors: | Kaur, S, Zhang, X.J, Schofield, C.J. | | Deposit date: | 2025-01-24 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Light-Induced, Lysine-Targeting Irreversible Covalent Inhibition of the Human Oxygen Sensing Hydroxylase Factor Inhibiting HIF (FIH).
J.Am.Chem.Soc., 147, 2025
|
|
4Y9J
 
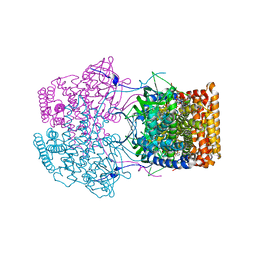 | |
6XAE
 
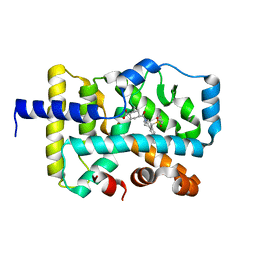 | | SUBSTITUTED BENZYLOXYTRICYCLIC COMPOUNDS AS RETINOIC ACID-RELATED ORPHAN RECEPTOR GAMMA T AGONISTS | | Descriptor: | Nuclear receptor ROR-gamma, trans-4-{(3aR,9bR)-7-[(2-chloro-6-fluorophenyl)methoxy]-9b-[(4-fluorophenyl)sulfonyl]-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl}cyclohexane-1-carboxylic acid | | Authors: | Sack, J.S. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.257 Å) | | Cite: | Substituted benzyloxytricyclic compounds as retinoic acid-related orphan receptor gamma t (ROR gamma t) agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4I1L
 
 | | Structural and Biological Features of FOXP3 Dimerization Relevant to Regulatory T Cell Function | | Descriptor: | ACETATE ION, Forkhead box protein P3, MAGNESIUM ION, ... | | Authors: | Song, X.M, Greene, M.I, Zhou, Z.C. | | Deposit date: | 2012-11-21 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function.
Cell Rep, 1, 2012
|
|
4Y9L
 
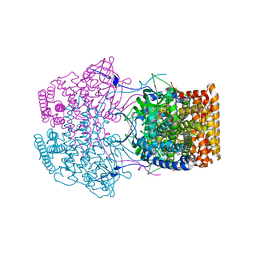 | | Crystal Structure of Caenorhabditis elegans ACDH-11 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Protein ACDH-11, isoform b | | Authors: | Li, Z.J, Zhai, Y.J, Zhang, K, Sun, F. | | Deposit date: | 2015-02-17 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Acyl-CoA Dehydrogenase Drives Heat Adaptation by Sequestering Fatty Acids
Cell, 161, 2015
|
|
9JH5
 
 | | Activation mechanism of CYSLTR2 by C16:0 ceramide | | Descriptor: | CHOLESTEROL, Cysteinyl leukotriene receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2024-09-09 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Sensing ceramides by CYSLTR2 and P2RY6 to aggravate atherosclerosis.
Nature, 641, 2025
|
|
9JH6
 
 | | Activation mechanism of CYSLTR2 by C20:0 | | Descriptor: | Cer(d18:0/20:0), Cysteinyl leukotriene receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, J.L, Sun, J.P, Yu, X. | | Deposit date: | 2024-09-09 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Sensing ceramides by CYSLTR2 and P2RY6 to aggravate atherosclerosis.
Nature, 641, 2025
|
|
6A0X
 
 | | Crystal structure of broadly neutralizing antibody 13D4 | | Descriptor: | Antibody 13D4, Fab Heavy Chain, Fab Light Chain | | Authors: | Li, S, Li, T. | | Deposit date: | 2018-06-06 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Broad, Antibody-Mediated Neutralization of H5N1 Influenza Virus.
J. Virol., 92, 2018
|
|
