2VJU
 
 | | Crystal structure of the IS608 transposase in complex with the complete Right end 35-mer DNA and manganese | | Descriptor: | MANGANESE (II) ION, RIGHT END 35-MER, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
1SHP
 
 | | THE NMR SOLUTION STRUCTURE OF A KUNITZ-TYPE PROTEINASE INHIBITOR FROM THE SEA ANEMONE STICHODACTYLA HELIANTHUS | | Descriptor: | TRYPSIN INHIBITOR | | Authors: | Antuch, W, Berndt, K, Chavez, M, Delfin, J, Wuthrich, K. | | Deposit date: | 1992-11-17 | | Release date: | 1994-01-31 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of a Kunitz-type proteinase inhibitor from the sea anemone Stichodactyla helianthus.
Eur.J.Biochem., 212, 1993
|
|
1SPY
 
 | | REGULATORY DOMAIN OF HUMAN CARDIAC TROPONIN C IN THE CALCIUM-FREE STATE, NMR, 40 STRUCTURES | | Descriptor: | TROPONIN C | | Authors: | Spyracopoulos, L, Li, M.X, Sia, S.K, Gagne, S.M, Chandra, M, Solaro, R.J, Sykes, B.D. | | Deposit date: | 1997-07-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural transition in the regulatory domain of human cardiac troponin C.
Biochemistry, 36, 1997
|
|
2LDD
 
 | | Solution structure of the estrogen receptor-binding stapled peptide SP6 (Ac-EKHKILXRLLXDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP6 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-21 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2LDA
 
 | | Solution structure of the estrogen receptor-binding stapled peptide SP2 (Ac-HKXLHQXLQDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP2 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
2K28
 
 | | Solution NMR structure of the chromo domain of the chromobox protein homolog 4 | | Descriptor: | E3 SUMO-protein ligase CBX4 | | Authors: | Kaustov, L, Lemak, A, Quyang, H, Fares, C, Gutmanas, A, Ravichandran, M, Loppnau, P, Bountra, C, Weigelt, J, Edwards, A.M, Min, J, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the chromo domain of the chromobox protein homolog 4.
To be Published
|
|
2K1B
 
 | | Solution NMR structure of the chromo domain of the chromobox protein homolog 7 | | Descriptor: | Chromobox protein homolog 7 | | Authors: | Kaustov, L, Lemak, A, Quyang, H, Gutmanas, A, Fares, C, Bountra, C, Weigelt, J, Loppnau, P, Ravichandran, M, Edwards, A.M, Min, J, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the chromobox protein homolog 7.
To be Published
|
|
2LDC
 
 | | Solution structure of the estrogen receptor-binding stapled peptide SP1 (Ac-HXILHXLLQDS-NH2) | | Descriptor: | Estrogen receptor-binding stapled peptide SP1 | | Authors: | Phillips, C, Bazin, R, Bent, A, Davies, N, Moore, R, Pannifer, A, Pickford, A, Prior, S, Read, C, Roberts, L, Schade, M, Scott, A, Brown, D, Xu, B, Irving, S. | | Deposit date: | 2011-05-20 | | Release date: | 2011-07-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Design and structure of stapled peptides binding to estrogen receptors.
J.Am.Chem.Soc., 133, 2011
|
|
6XOG
 
 | | Structure of SUMO1-ML786519 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOH
 
 | | Structure of SUMO1-ML00789344 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOI
 
 | | Structure of SUMO1-ML00752641 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
8B3E
 
 | | Variant Surface Glycoprotein VSG397 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Variant surface glycoprotein 397 | | Authors: | Zeelen, J.P, Stebbins, C.E, Dakovic, S, Foti, K. | | Deposit date: | 2022-09-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural similarities between the metacyclic and bloodstream form variant surface glycoproteins of the African trypanosome.
Plos Negl Trop Dis, 17, 2023
|
|
6T3O
 
 | | Crystal structure of the human myomesin domain 10 | | Descriptor: | AZIDE ION, Myomesin-1 | | Authors: | Duskova, J, Petrokova, H, Maly, P. | | Deposit date: | 2019-10-11 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Myomedin scaffold variants targeted to 10E8 HIV-1 broadly neutralizing antibody mimic gp41 epitope and elicit HIV-1 virus-neutralizing sera in mice.
Virulence, 12, 2021
|
|
7NE3
 
 | | Human TET2 in complex with favourable DNA substrate. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*CP*AP*GP*GP*(5CM)P*GP*CP*CP*TP*G)-3'), ... | | Authors: | Rafalski, D, Bochtler, M. | | Deposit date: | 2021-02-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Pronounced sequence specificity of the TET enzyme catalytic domain guides its cellular function.
Sci Adv, 8, 2022
|
|
7NE6
 
 | | Human TET2 in complex with unfavourable DNA substrate. | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*AP*CP*AP*GP*GP*(5CM)P*GP*CP*CP*TP*G)-3'), ... | | Authors: | Rafalski, D, Bochtler, M. | | Deposit date: | 2021-02-03 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pronounced sequence specificity of the TET enzyme catalytic domain guides its cellular function.
Sci Adv, 8, 2022
|
|
5LSF
 
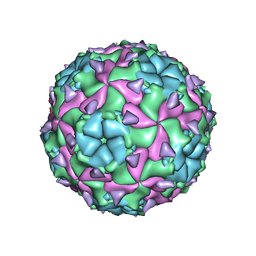 | | Sacbrood honeybee virus | | Descriptor: | CALCIUM ION, VP1, VP2, ... | | Authors: | Plevka, P, Veselikova, M. | | Deposit date: | 2016-08-26 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Virion structure and genome delivery mechanism of sacbrood honeybee virus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7MZV
 
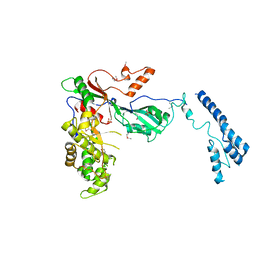 | | Structure of yeast pseudouridine synthase 7 (PUS7) | | Descriptor: | Multisubstrate pseudouridine synthase 7, SULFATE ION | | Authors: | Purchal, M, Koutmos, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Pseudouridine synthase 7 is an opportunistic enzyme that binds and modifies substrates with diverse sequences and structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5WOE
 
 | |
5UKE
 
 | |
5G1X
 
 | | Crystal structure of Aurora-A kinase in complex with N-Myc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AURORA KINASE A, MAGNESIUM ION, ... | | Authors: | Richards, M.W, Burgess, S.G, Bayliss, R. | | Deposit date: | 2016-03-31 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Basis of N-Myc Binding by Aurora-A and its Destabilization by Kinase Inhibitors
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5C7G
 
 | |
1EGZ
 
 | | CELLULASE CEL5 FROM ERWINIA CHRYSANTHEMI, A FAMILY GH 5-2 ENZYME | | Descriptor: | CALCIUM ION, ENDOGLUCANASE Z | | Authors: | Czjzek, M, El Hassouni, M, Py, B, Barras, F. | | Deposit date: | 1999-03-18 | | Release date: | 1999-03-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Type II protein secretion in gram-negative pathogenic bacteria: the study of the structure/secretion relationships of the cellulase Cel5 (formerly EGZ) from Erwinia chrysanthemi
J.Mol.Biol., 310, 2001
|
|
4K3J
 
 | | Crystal structure of Onartuzumab Fab in complex with MET and HGF-beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor beta chain, ... | | Authors: | Ma, X, Starovasnik, M.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Monovalent antibody design and mechanism of action of onartuzumab, a MET antagonist with anti-tumor activity as a therapeutic agent.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1SHO
 
 | |
5UTF
 
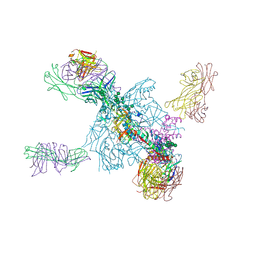 | | Crystal Structure of a Stabilized DS-SOSIP.6mut BG505 gp140 HIV-1 Env Trimer, Containing Mutations I201C-P433C (DS), L154M, Y177W, N300M, N302M, T320L, I420M in Complex with Human Antibodies PGT122 and 35O22 at 4.3 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35022 Heavy chain, ... | | Authors: | Pancera, M, Chuang, G.-Y, Xu, K, Kwong, P.D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity.
J. Virol., 91, 2017
|
|
