8H82
 
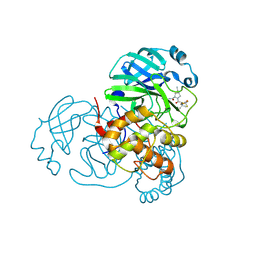 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (E166V) in complex with protease inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-10-21 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8HBK
 
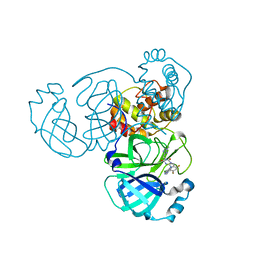 | | The crystal structure of SARS-CoV-2 3CL protease in complex with Ensitrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Lin, M. | | Deposit date: | 2022-10-29 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H4Y
 
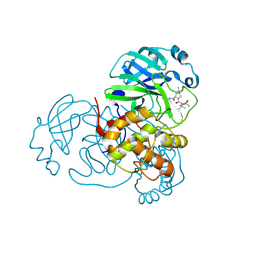 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) F140L Mutant in Complex with Inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lin, M, Liu, X. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
3PUR
 
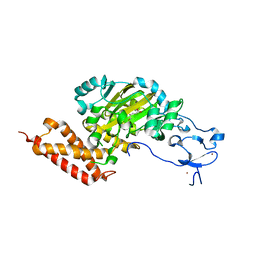 | | CEKDM7A from C.Elegans, complex with D-2-HG | | Descriptor: | (2R)-2-hydroxypentanedioic acid, FE (II) ION, Lysine-specific demethylase 7 homolog, ... | | Authors: | Yang, Y, Wang, P, Xu, W, Xu, Y. | | Deposit date: | 2010-12-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
3PUQ
 
 | | CEKDM7A from C.Elegans, complex with alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, GLYCEROL, ... | | Authors: | Yang, Y, Wang, P, Xu, W, Xu, Y. | | Deposit date: | 2010-12-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
8GOU
 
 | | Omicron BA.4/5 SARS-CoV-2 S in complex with TH003 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, TH003 Fab heavy chain, ... | | Authors: | Guo, Y, Zhang, G, Liang, J, Liu, F, Rao, Z. | | Deposit date: | 2022-08-25 | | Release date: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
8GPY
 
 | | Crystal structure of Omicron BA.4/5 RBD in complex with a neutralizing antibody scFv | | Descriptor: | Spike protein S1, scFv | | Authors: | Gao, Y.X, Song, Z.D, Wang, W.M, Guo, Y. | | Deposit date: | 2022-08-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and characterization of potent pan-variant SARS-CoV-2 neutralizing antibodies from individuals with Omicron breakthrough infection.
Nat Commun, 14, 2023
|
|
4B4S
 
 | |
4C5D
 
 | | Crystal structure of Bcl-xL in complex with benzoylurea compound (42) | | Descriptor: | (R)-3-(4-BROMOBENZYLTHIO)-2-(3-(3-((2,4-DIFLUOROPHENYL)ETHYNYL)BENZOYL)-3-PROPYLUREIDO)PROPANOIC ACID, 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Roy, M.J, Brady, R.M, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De-Novo Designed Library of Benzoylureas as Inhibitors of Bcl-Xl: Synthesis, Structural and Biochemical Characterization.
J.Med.Chem., 57, 2014
|
|
8K11
 
 | | SID1 transmembrane family member 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K12
 
 | | SID1 transmembrane family member 2 | | Descriptor: | SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K1B
 
 | | SID1 transmembrane family member 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K10
 
 | | SID1 transmembrane family member 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K13
 
 | | SID1 transmembrane family member 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K1D
 
 | | SID1 transmembrane family member 1 | | Descriptor: | SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
4C52
 
 | | Crystal structure of Bcl-xL in complex with benzoylurea compound (39b) | | Descriptor: | (R)-2-(3-(3-((2,4-DIFLUOROPENYL)ETHYNYL)BENZOYL)-3-PROPYLUREIDO)-3-(ISOBUTYLTHIO) PROPANOIC ACID, 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Roy, M.J, Brady, R.M, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2013-09-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | De-Novo Designed Library of Benzoylureas as Inhibitors of Bcl-Xl: Synthesis, Structural and Biochemical Characterization.
J.Med.Chem., 57, 2014
|
|
3ZLO
 
 | | Crystal structure of BCL-XL in complex with inhibitor (Compound 6) | | Descriptor: | 2-[(8E)-8-(1,3-benzothiazol-2-ylhydrazinylidene)-6,7-dihydro-5H-naphthalen-2-yl]-5-(4-phenylbutyl)-1,3-thiazole-4-carboxylic acid, BCL-2-LIKE PROTEIN 1 | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-02-04 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
3ZLR
 
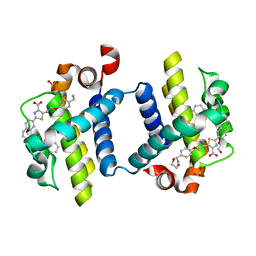 | | Crystal structure of BCL-XL in complex with inhibitor (WEHI-539) | | Descriptor: | 1,2-ETHANEDIOL, 5-[3-[4-(aminomethyl)phenoxy]propyl]-2-[(8E)-8-(1,3-benzothiazol-2-ylhydrazinylidene)-6,7-dihydro-5H-naphthalen-2-yl]-1,3-thiazole-4-carboxylic acid, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-02-04 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
4CI2
 
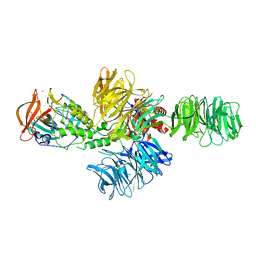 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to lenalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Lenalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
3ZLN
 
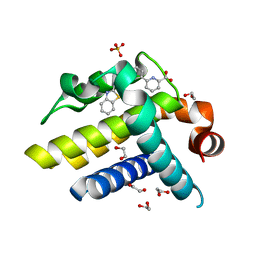 | | Crystal structure of BCL-XL in complex with inhibitor (Compound 3) | | Descriptor: | 1,2-ETHANEDIOL, 6-[(8E)-8-(1,3-benzothiazol-2-ylhydrazinylidene)-6,7-dihydro-5H-naphthalen-2-yl]pyridine-2-carboxylic acid, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-02-04 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
3ZK6
 
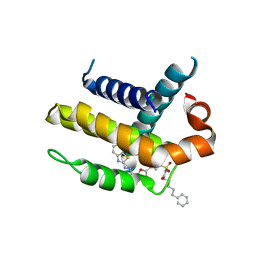 | | Crystal structure of Bcl-xL in complex with inhibitor (Compound 2). | | Descriptor: | BCL-2-LIKE PROTEIN 1, N-(3-(5-(1-(2-(benzo[d]thiazol-2-yl)hydrazono)ethyl)furan-2-yl)phenylsulfonyl)-6-phenylhexanamide | | Authors: | Czabotar, P.E, Lessene, G.L, Smith, B.J, Colman, P.M. | | Deposit date: | 2013-01-22 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure-Guided Design of a Selective Bcl-Xl Inhibitor
Nat.Chem.Biol., 9, 2013
|
|
4CI3
 
 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to Pomalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Pomalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4CI1
 
 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to thalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Thalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
4P6W
 
 | | Crystal Structure of mometasone furoate-bound glucocorticoid receptor ligand binding domain | | Descriptor: | Glucocorticoid receptor, MOMETASONE FUROATE, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
4P6X
 
 | | Crystal Structure of cortisol-bound glucocorticoid receptor ligand binding domain | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
