8CN3
 
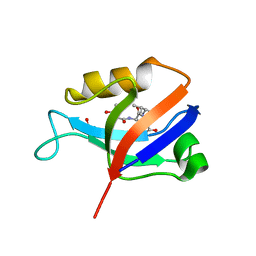 | | hDLG1-PDZ2 in complex with a TAX1 peptide from HTLV-1 | | Descriptor: | Disks large homolog 1, GLU-THR-GLU-VAL | | Authors: | Maseko, S, Sogues, A, Volkov, A, Remaut, H, Twizere, J.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Identification of small molecule antivirals against HTLV-1 by targeting the hDLG1-Tax-1 protein-protein interaction.
Antiviral Res., 217, 2023
|
|
8CN1
 
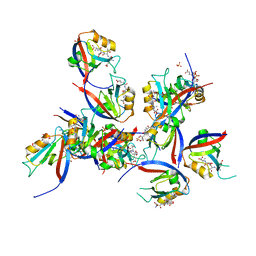 | | hDLG1-PDZ1 in complex with a TAX1 peptide from HTLV-1 | | Descriptor: | Disks large homolog 1, GLU-THR-GLU-VAL, SULFATE ION | | Authors: | Maseko, S, Sogues, A, Volkov, A, Remaut, H, Twizere, J.C. | | Deposit date: | 2023-02-21 | | Release date: | 2023-08-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Identification of small molecule antivirals against HTLV-1 by targeting the hDLG1-Tax-1 protein-protein interaction.
Antiviral Res., 217, 2023
|
|
8CB7
 
 | |
8CBG
 
 | |
8CB5
 
 | |
8CB4
 
 | |
8CBP
 
 | |
8CBI
 
 | |
5IVQ
 
 | |
5JAQ
 
 | | Yersinia pestis FabV variant T276C | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia pestis FabV Enoyl-ACP Reductase.
Biochemistry, 55, 2016
|
|
5LWO
 
 | | Structure of Spin-labelled T4 lysozyme mutant L115C-R119C-R1 at 100K | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Loll, B, Consentius, P, Gohlke, U, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | Deposit date: | 2016-09-18 | | Release date: | 2017-03-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | Internal Dynamics of the 3-Pyrroline-N-Oxide Ring in Spin-Labeled Proteins.
J Phys Chem Lett, 8, 2017
|
|
6T4A
 
 | | Thrombin in Complex with a D-Phe-Pro-p-aminopyridine derivative | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(6-azanylpyridin-3-yl)methyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Ngo, K, Collins, C, Heine, A, Klebe, G. | | Deposit date: | 2019-10-13 | | Release date: | 2020-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Protein-Induced Change in Ligand Protonation during Trypsin and Thrombin Binding: Hint on Differences in Selectivity Determinants of Both Proteins?
J.Med.Chem., 63, 2020
|
|
6T3Q
 
 | | Thrombin in Complex with a D-Phe-Pro-2-aminopyridine derivative | | Descriptor: | (2~{S})-1-[(2~{R})-2-azanyl-3-phenyl-propanoyl]-~{N}-[(2-azanylpyridin-4-yl)methyl]pyrrolidine-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Ngo, K, Collins, C, Heine, A, Klebe, G. | | Deposit date: | 2019-10-11 | | Release date: | 2020-05-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Protein-Induced Change in Ligand Protonation during Trypsin and Thrombin Binding: Hint on Differences in Selectivity Determinants of Both Proteins?
J.Med.Chem., 63, 2020
|
|
6WKY
 
 | | Cryo-EM of Form 1 related peptide filament, 29-24-3 | | Descriptor: | peptide 29-24-3 | | Authors: | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WPE
 
 | | HUMAN IDO1 IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 4-chloro-N-{[1-(3-chlorobenzene-1-carbonyl)-1,2,3,4-tetrahydroquinolin-6-yl]methyl}benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lesburg, C.A, Lammens, A. | | Deposit date: | 2020-04-27 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Carbamate and N -Pyrimidine Mitigate Amide Hydrolysis: Structure-Based Drug Design of Tetrahydroquinoline IDO1 Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
5K3Y
 
 | | Crystal structure of AuroraB/INCENP in complex with BI 811283 | | Descriptor: | Aurora kinase B-A, Inner centromere protein A, N-methyl-N-(1-methylpiperidin-4-yl)-4-{[4-({(1R,2S)-2-[(propan-2-yl)carbamoyl]cyclopentyl}amino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}benzamide | | Authors: | Bader, G, Zahn, S.K, Zoephel, A. | | Deposit date: | 2016-05-20 | | Release date: | 2016-08-17 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pharmacological Profile of BI 847325, an Orally Bioavailable, ATP-Competitive Inhibitor of MEK and Aurora Kinases.
Mol.Cancer Ther., 15, 2016
|
|
8A53
 
 | | Crystal structure of AtMCA-IIf C147A (metacaspase 9) from Arabidopsis thaliana | | Descriptor: | Metacaspase-9, NITRATE ION | | Authors: | Sabljic, I, Stael, S, Stahlberg, J, Bozhkov, P. | | Deposit date: | 2022-06-14 | | Release date: | 2023-05-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function study of a Ca 2+ -independent metacaspase involved in lateral root emergence.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5KML
 
 | |
5KMN
 
 | |
5KMK
 
 | |
5KMO
 
 | | TrkA JM-kinase with 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-(2-pyridyl)urea | | Descriptor: | 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-pyridin-2-yl-urea, High affinity nerve growth factor receptor | | Authors: | Su, H.P. | | Deposit date: | 2016-06-27 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural characterization of nonactive site, TrkA-selective kinase inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KMI
 
 | |
5KMM
 
 | | TrkA JM-kinase with 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-(1-naphthyl)urea | | Descriptor: | 1-(2-methyl-4-phenyl-pyrimidin-5-yl)-3-naphthalen-1-yl-urea, High affinity nerve growth factor receptor | | Authors: | Su, H.P. | | Deposit date: | 2016-06-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural characterization of nonactive site, TrkA-selective kinase inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KMJ
 
 | | TrkA JM-kinase with {N}-(2-pyridylmethyl)-2-[2-(2-thienyl)indol-1-yl]acetamide | | Descriptor: | High affinity nerve growth factor receptor, ~{N}-(pyridin-2-ylmethyl)-2-(2-thiophen-2-ylindol-1-yl)ethanamide | | Authors: | Su, H.P. | | Deposit date: | 2016-06-27 | | Release date: | 2016-12-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural characterization of nonactive site, TrkA-selective kinase inhibitors.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8DYA
 
 | |
