8GW1
 
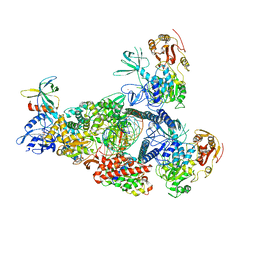 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | Helicase, MANGANESE (II) ION, Non-structural protein 7, ... | | Authors: | Yan, L, Rao, Z, Lou, Z. | | Deposit date: | 2022-09-16 | | Release date: | 2023-10-25 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWM
 
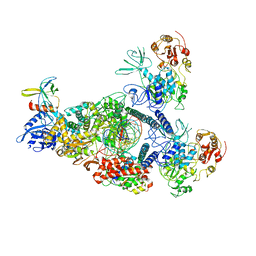 | | SARS-CoV-2 E-RTC bound with MMP-nsp9 and GMPPNP | | Descriptor: | 2'-deoxy-2'-fluoro-2'-methyluridine 5'-(trihydrogen diphosphate), Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
3B4D
 
 | | Crystal Structure of Human PABPN1 RRM | | Descriptor: | Polyadenylate-binding protein 2 | | Authors: | Ge, H, Tong, S, Teng, M, Niu, L. | | Deposit date: | 2007-10-24 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure and possible dimerization of the single RRM of human PABPN1
Proteins, 71, 2008
|
|
1PVD
 
 | |
3B9C
 
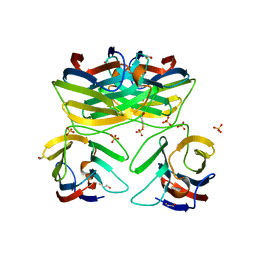 | | Crystal Structure of Human GRP CRD | | Descriptor: | BETA-MERCAPTOETHANOL, HSPC159, SULFATE ION | | Authors: | Zhou, D, Ge, H.H, Niu, L.W, Teng, M.K. | | Deposit date: | 2007-11-05 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the C-terminal conserved domain of human GRP, a galectin-related protein, reveals a function mode different from those of galectins.
Proteins, 71, 2008
|
|
3B4M
 
 | | Crystal Structure of Human PABPN1 RRM | | Descriptor: | Polyadenylate-binding protein 2 | | Authors: | Ge, H, Zhou, D, Teng, M, Niu, L. | | Deposit date: | 2007-10-24 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure and possible dimerization of the single RRM of human PABPN1
Proteins, 71, 2008
|
|
3BS8
 
 | | Crystal structure of Glutamate 1-Semialdehyde Aminotransferase complexed with pyridoxamine-5'-phosphate From Bacillus subtilis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Glutamate-1-semialdehyde 2,1-aminomutase | | Authors: | Ge, H, Fan, J, Teng, M, Niu, L. | | Deposit date: | 2007-12-22 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Glutamate1-semialdehyde aminotransferase from Bacillus subtilis with bound pyridoxamine-5'-phosphate
Biochem.Biophys.Res.Commun., 402, 2010
|
|
7F3U
 
 | | Cryo-EM structure of human TMEM120A in the CoASH-free state | | Descriptor: | Transmembrane protein 120A | | Authors: | Rong, Y, Gao, Y.W, Song, D.F, Zhao, Y, Liu, Z.F. | | Deposit date: | 2021-06-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | TMEM120A contains a specific coenzyme A-binding site and might not mediate poking- or stretch-induced channel activities in cells.
Elife, 10, 2021
|
|
8H6T
 
 | | Complex structure of CDK2/Cyclin E1 and a potent, selective small molecule inhibitor | | Descriptor: | (1R,3S)-3-{3-[(pyridin-2-yl)amino]-1H-pyrazol-5-yl}cyclopentyl propan-2-ylcarbamate, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
8H6P
 
 | | Complex structure of CDK2/Cyclin E1 and a potent, selective macrocyclic inhibitor | | Descriptor: | (7S,10R)-11-oxa-2,4,5,13,17,23-hexaazatetracyclo[17.3.1.1~3,6~.1~7,10~]pentacosa-1(23),3(25),5,19,21-pentaene-12,18-dione, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Ren, X. | | Deposit date: | 2022-10-18 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design.
Acs Med.Chem.Lett., 14, 2023
|
|
4LUQ
 
 | |
4XVM
 
 | |
4XVI
 
 | |
8HFK
 
 | |
8HFJ
 
 | | Crystal Structure of CbAR mutant (H162F) in complex with NADP+ and a bulky 1,3-cyclodiketone | | Descriptor: | 2-methyl-2-[(4-methylphenyl)methyl]cyclopentane-1,3-dione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Versicolorin reductase | | Authors: | Hou, X.D, Yin, D.J, Rao, Y.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of an anthrol reductase inspires enantioselective synthesis of enantiopure hydroxycycloketones and beta-halohydrins.
Nat Commun, 14, 2023
|
|
4XVK
 
 | | Binary complex of human polymerase nu and DNA with the finger domain closed | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*GP*AP*TP*CP*TP*GP*AP*CP*GP*CP*TP*AP*CP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*GP*TP*AP*GP*CP*GP*TP*CP*A)-3'), ... | | Authors: | Lee, Y.-S, Yang, W. | | Deposit date: | 2015-01-27 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | How a homolog of high-fidelity replicases conducts mutagenic DNA synthesis.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4XVL
 
 | | Binary complex of human polymerase nu and DNA with the finger domain open | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA (5'-D(*GP*AP*TP*CP*TP*GP*AP*CP*GP*CP*TP*AP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*TP*AP*GP*CP*GP*TP*CP*A)-3'), ... | | Authors: | Lee, Y.-S, Yang, W. | | Deposit date: | 2015-01-27 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | How a homolog of high-fidelity replicases conducts mutagenic DNA synthesis.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2MBE
 
 | |
3MIL
 
 | | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Isoamyl acetate-hydrolyzing esterase | | Authors: | Ma, J, Lu, Q, Yuan, Y, Li, K, Ge, H, Go, Y, Niu, L, Teng, M. | | Deposit date: | 2010-04-11 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae reveals a novel active site architecture and the basis of substrate specificity
Proteins, 79, 2011
|
|
7FBT
 
 | | Crystal structure of chitinase (RmChi1) from Rhizomucor miehei (sp p32 2 1, MR) | | Descriptor: | Chitinase, MAGNESIUM ION | | Authors: | Jiang, Z.Q, Hu, S.Q, Zhu, Q, Liu, Y.C, Ma, J.W, Yan, Q.J, Gao, Y.G, Yang, S.Q. | | Deposit date: | 2021-07-12 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a chitinase (RmChiA) from the thermophilic fungus Rhizomucor miehei with a real active site tunnel.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
7TIL
 
 | | CryoEM structure of JetD from Pseudomonas aeruginosa | | Descriptor: | JetD | | Authors: | Deep, A, Gu, Y, Gao, Y, Ego, K, Herzik, M, Zhou, H, Corbett, K. | | Deposit date: | 2022-01-13 | | Release date: | 2022-10-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The SMC-family Wadjet complex protects bacteria from plasmid transformation by recognition and cleavage of closed-circular DNA.
Mol.Cell, 82, 2022
|
|
2ID5
 
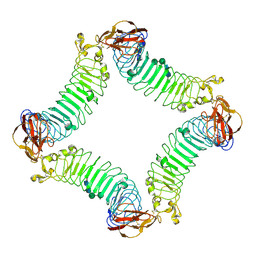 | | Crystal Structure of the Lingo-1 Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine rich repeat neuronal 6A, ... | | Authors: | Mosyak, L, Wood, A, Dwyer, B, Johnson, M, Stahl, M.L, Somers, W.S. | | Deposit date: | 2006-09-14 | | Release date: | 2006-09-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition.
J.Biol.Chem., 281, 2006
|
|
3JBZ
 
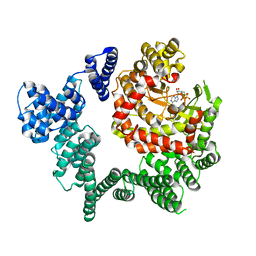 | |
7MTQ
 
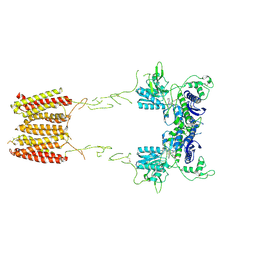 | | CryoEM Structure of Full-Length mGlu2 in Inactive-State Bound to Antagonist LY341495 | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Seven, A.B, Barros-Alvarez, X, Skiniotis, G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | G-protein activation by a metabotropic glutamate receptor.
Nature, 595, 2021
|
|
7MTR
 
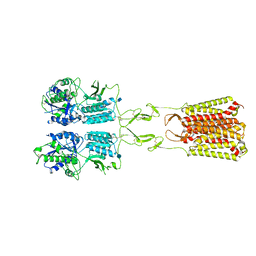 | | CryoEM Structure of Full-Length mGlu2 Bound to Ago-PAM ADX55164 and Glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methoxy-6-propyl-N-(2-{4-[(1H-tetrazol-5-yl)methoxy]phenyl}ethyl)thieno[2,3-d]pyrimidin-4-amine, GLUTAMIC ACID, ... | | Authors: | Seven, A.B, Barros-Alvarez, X, Skiniotis, G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-09-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | G-protein activation by a metabotropic glutamate receptor.
Nature, 595, 2021
|
|
