7RXJ
 
 | |
7S0X
 
 | |
6BL0
 
 | | Novel Modes of Inhibition of Wild-Type IDH1:Direct Covalent Modification of His315 with Cmpd11 | | Descriptor: | (5aS,6S,8S,9aS)-2-(benzenecarbonyl)-6-methyl-7-oxo-9a-phenyl-4,5,5a,6,7,8,9,9a-octahydro-2H-benzo[g]indazole-8-carbonitrile, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2017-11-09 | | Release date: | 2018-07-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
7PON
 
 | |
7PNO
 
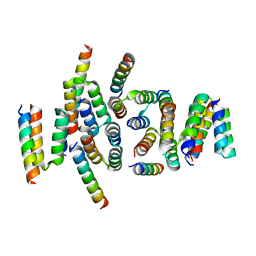 | | C terminal domain of Nipah Virus Phosphoprotein fused to the Ntail alpha more of the Nucleoprotein. | | Descriptor: | Phosphoprotein, alpha MoRE of Nipah virus Nucleoprotein tail | | Authors: | Bourhis, J.M, Yabukaski, F, Tarbouriech, N, Jamin, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Dynamics of the C-terminal X Domain of Nipah and Hendra Viruses Controls the Attachment to the C-terminal Tail of the Nucleocapsid Protein.
J.Mol.Biol., 434, 2022
|
|
6MSC
 
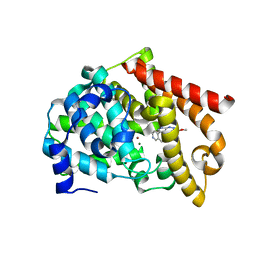 | | Novel, potent, selective and brain penetrant phosphodiesterase 10A inhibitors | | Descriptor: | 8-fluoro-6-methoxy-3-methyl-1-(3-methylpyridin-4-yl)-3H-pyrazolo[3,4-c]cinnoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Jakob, C.G. | | Deposit date: | 2018-10-16 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Novel, potent, selective, and brain penetrant phosphodiesterase 10A inhibitors.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
5NIQ
 
 | |
8BXL
 
 | | Patulin Synthase from Penicillium expansum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tjallinks, G, Boverio, A, Rozeboom, H.J, Fraaije, M.W. | | Deposit date: | 2022-12-09 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure elucidation and characterization of patulin synthase, insights into the formation of a fungal mycotoxin.
Febs J., 290, 2023
|
|
8BZQ
 
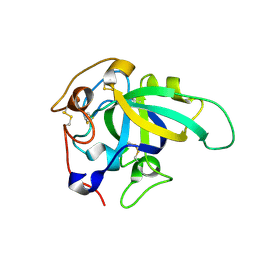 | |
5I04
 
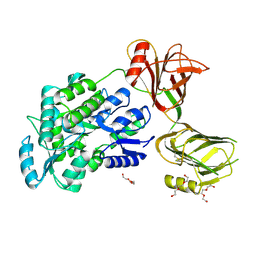 | | Crystal structure of the orphan region of human endoglin/CD105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Endoglin, TRIETHYLENE GLYCOL, ... | | Authors: | Saito, T, Bokhove, M, de Sanctis, D, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
7ZAO
 
 | | Structure of BPP43_05035 of Brachyspira pilosicoli | | Descriptor: | MAGNESIUM ION, Sialidase (Neuraminidase) family protein-like protein | | Authors: | Rajan, A, Gallego, P, Pelaseyed, T. | | Deposit date: | 2022-03-22 | | Release date: | 2023-10-11 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | BPP43_05035 is a Brachyspira pilosicoli cell surface adhesin that weakens the integrity of the epithelial barrier during infection
Biorxiv, 2024
|
|
6MSA
 
 | | Novel, potent, selective and brain penetrant phosphodiesterase 10A inhibitors | | Descriptor: | 7,8-dimethoxy-1-methyl-2H-pyrazolo[3,4-c]cinnoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Jakob, C.G. | | Deposit date: | 2018-10-16 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Novel, potent, selective, and brain penetrant phosphodiesterase 10A inhibitors.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
9AV0
 
 | | Crystal structure of S. aureus GuaB dCBS with inhibitor GNE2011 | | Descriptor: | 9-{(1R)-1-[(5P)-5-(4-chloro-1H-imidazol-2-yl)pyridin-3-yl]ethoxy}-1,4-dihydro-2H-pyrano[3,4-c]quinoline, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2024-11-27 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
5JH3
 
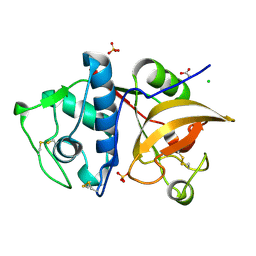 | | Human cathepsin K mutant C25S | | Descriptor: | ACETATE ION, CHLORIDE ION, Cathepsin K, ... | | Authors: | Novinec, M, Korenc, M, Lenarcic, B. | | Deposit date: | 2016-04-20 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An allosteric site enables fine-tuning of cathepsin K by diverse effectors.
FEBS Lett., 590, 2016
|
|
6DSO
 
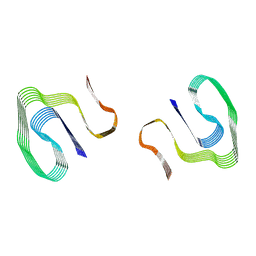 | | Cryo-EM structure of murine AA amyloid fibril | | Descriptor: | Serum amyloid A-2 protein | | Authors: | Loerch, S, Liberta, F, Grigorieff, N, Fandrich, M, Schmidt, M. | | Deposit date: | 2018-06-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids.
Nat Commun, 10, 2019
|
|
9FCT
 
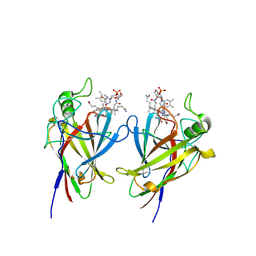 | |
9AUW
 
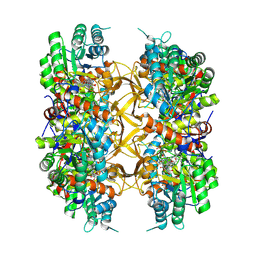 | | Crystal structure of A. baumannii GuaB dCBS with inhibitor GNE9979 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-[4-chloro-3-(dimethylamino)phenyl]-N~2~-[3-(hydroxymethyl)quinolin-6-yl]-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUV
 
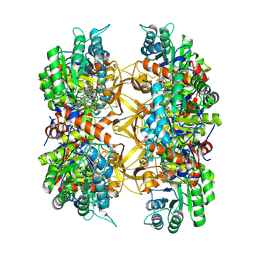 | | Crystal structure of A. baumannii GuaB dCBS with inhibitor GNE9123 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(6-chloropyridin-3-yl)-N~2~-(1,4-dihydro-2H-pyrano[3,4-c]quinolin-9-yl)-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUX
 
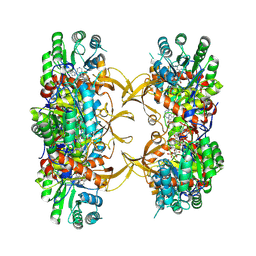 | | Crystal structure of A. baumannii GuaB dCBS with inhibitor GNE2011 | | Descriptor: | 9-{(1R)-1-[(5P)-5-(4-chloro-1H-imidazol-2-yl)pyridin-3-yl]ethoxy}-1,4-dihydro-2H-pyrano[3,4-c]quinoline, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUZ
 
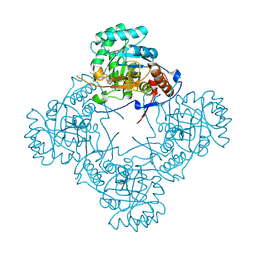 | | Crystal structure of S. aureus GuaB dCBS with inhibitor GNE9979 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-[4-chloro-3-(dimethylamino)phenyl]-N~2~-[3-(hydroxymethyl)quinolin-6-yl]-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AV2
 
 | | Crystal structure of E. coli GuaB dCBS with inhibitor GNE9979 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-[4-chloro-3-(dimethylamino)phenyl]-N~2~-[3-(hydroxymethyl)quinolin-6-yl]-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AUY
 
 | | Crystal structure of S. aureus GuaB dCBS with inhibitor GNE9123 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(6-chloropyridin-3-yl)-N~2~-(1,4-dihydro-2H-pyrano[3,4-c]quinolin-9-yl)-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AV1
 
 | | Crystal structure of E. coli GuaB dCBS with inhibitor GNE9123 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-(6-chloropyridin-3-yl)-N~2~-(1,4-dihydro-2H-pyrano[3,4-c]quinolin-9-yl)-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
9AV3
 
 | | Crystal structure of E. coli GuaB dCBS with inhibitor GNE2011 | | Descriptor: | 9-{(1R)-1-[(5P)-5-(4-chloro-1H-imidazol-2-yl)pyridin-3-yl]ethoxy}-1,4-dihydro-2H-pyrano[3,4-c]quinoline, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-03-01 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of GuaB inhibitors with efficacy against Acinetobacter baumannii infection.
Mbio, 15, 2024
|
|
7NAB
 
 | | Crystal structure of human neutralizing mAb CV3-25 binding to SARS-CoV-2 S MPER peptide 1140-1165 | | Descriptor: | CITRIC ACID, CV3-25 Fab Heavy Chain, CV3-25 Fab Light Chain, ... | | Authors: | Chen, Y, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2021-06-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern.
Cell Rep, 38, 2022
|
|
