2GQM
 
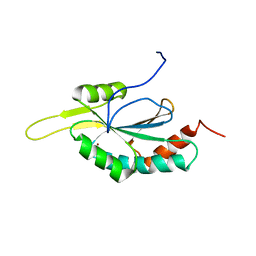 | | Solution structure of Human Cu(I)-Sco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3IZJ
 
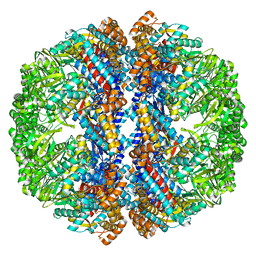 | | Mm-cpn rls with ATP and AlFx | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
2GQL
 
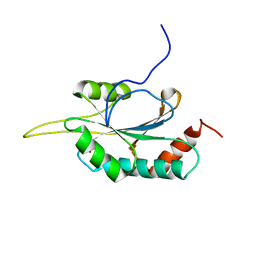 | | Solution structure of Human Ni(II)-Sco1 | | Descriptor: | NICKEL (II) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-21 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3N30
 
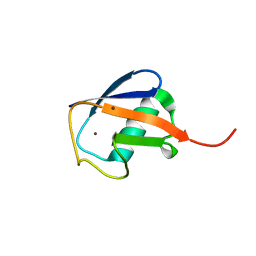 | | Crystal Structure of cubic Zn3-hUb (human ubiquitin) adduct | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Siliqi, D, Caliandro, R, Arnesano, F, Natile, G, Falini, G, Fermani, S, Belviso, B.D. | | Deposit date: | 2010-05-19 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic analysis of metal-ion binding to human ubiquitin.
Chemistry, 17, 2011
|
|
3LOS
 
 | | Atomic Model of Mm-cpn in the Closed State | | Descriptor: | Chaperonin | | Authors: | Zhang, J, Baker, M.L, Schroeder, G, Douglas, N.R, Reissmann, S, Jakana, J, Dougherty, M, Fu, C.J, Levitt, M, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2010-02-04 | | Release date: | 2010-03-16 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of folding chamber closure in a group II chaperonin
Nature, 463, 2010
|
|
6ZXO
 
 | |
3N32
 
 | | The crystal structure of human Ubiquitin adduct with Zeise's salt | | Descriptor: | PLATINUM (II) ION, Ubiquitin | | Authors: | Siliqi, D, Caliandro, R, Falini, G, Fermani, S, Natile, G, Arnesano, F, Belviso, B.D. | | Deposit date: | 2010-05-19 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Crystallographic analysis of metal-ion binding to human ubiquitin.
Chemistry, 17, 2011
|
|
6HF5
 
 | | Crystal Structure of the Acquired VIM-2 Metallo-beta-Lactamase in Complex with ANT-431 Inhibitor | | Descriptor: | 5-(pyridin-3-ylsulfonylamino)-1,3-thiazole-4-carboxylic acid, ACETATE ION, Beta-lactamase class B VIM-2, ... | | Authors: | Docquier, J.D, Pozzi, C, De Luca, F, Benvenuti, M, Mangani, S. | | Deposit date: | 2018-08-21 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | SAR Studies Leading to the Identification of a Novel Series of Metallo-beta-lactamase Inhibitors for the Treatment of Carbapenem-Resistant Enterobacteriaceae Infections That Display Efficacy in an Animal Infection Model.
Acs Infect Dis., 5, 2019
|
|
3U4R
 
 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-N-({3-[(methylsulfonyl)amino]phenyl}sulfonyl)-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxamide, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannagrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3U4O
 
 | | Novel HCV NS5B polymerase Inhibitors: Discovery of Indole C2 Acyl sulfonamides | | Descriptor: | 1-[(2-aminopyridin-4-yl)methyl]-5-chloro-3-(2-oxo-1,2-dihydropyridin-3-yl)-1H-indole-2-carboxylic acid, PHOSPHATE ION, RNA-directed RNA polymerase | | Authors: | Anilkumar, G.N, Selyutin, O, Rosenblum, S.B, Zeng, Q, Jiang, Y, Chan, T.-Y, Pu, H, Wang, L, Bennett, F, Chen, K.X, Lesburg, C.A, Duca, J.S, Gavalas, S, Huang, Y, Pinto, P, Sannigrahi, M, Velazquez, F, Venkataraman, S, Vilbubhan, B, Agrawal, S, Ferrari, E, Jiang, C.-K, Huang, H.-C, Shih, N.-Y, Njoroge, F.G, Kozlowski, J.A. | | Deposit date: | 2011-10-10 | | Release date: | 2011-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | II. Novel HCV NS5B polymerase inhibitors: Discovery of indole C2 acyl sulfonamides.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2YII
 
 | | Manipulating the regioselectivity of phenylalanine aminomutase: new insights into the reaction mechanism of MIO-dependent enzymes from structure-guided directed evolution | | Descriptor: | BETA-MERCAPTOETHANOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Wu, B, Szymanski, W, Wybenga, G.G, Heberling, M.M, Bartsch, S, Wildeman, S, Poelarends, G.J, Feringa, B.L, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Mechanism-Inspired Engineering of Phenylalanine Aminomutase for Enhanced Beta-Regioselective Asymmetric Amination of Cinnamates.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3P43
 
 | | Structure and Activities of Archaeal Members of the LigD 3' Phosphoesterase DNA Repair Enzyme Superfamily | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Smith, P, Nair, P.A, Das, U, Shuman, S. | | Deposit date: | 2010-10-05 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures and activities of archaeal members of the LigD 3'-phosphoesterase DNA repair enzyme superfamily.
Nucleic Acids Res., 39, 2011
|
|
2GVP
 
 | | Solution structure of Human apo Sco1 | | Descriptor: | SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Paulmaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GT5
 
 | | Solution structure of apo Human Sco1 | | Descriptor: | SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-27 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GT6
 
 | | Solution structure of Human Cu(I) Sco1 | | Descriptor: | COPPER (I) ION, SCO1 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mangani, S, Palumaa, P, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-04-27 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A hint for the function of human Sco1 from different structures.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1NBO
 
 | | The dual coenzyme specificity of photosynthetic glyceraldehyde-3-phosphate dehydrogenase interpreted by the crystal structure of A4 isoform complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, glyceraldehyde-3-phosphate dehydrogenase A | | Authors: | Falini, G, Fermani, S, Ripamonti, A, Sabatino, P, Sparla, F, Pupillo, P, Trost, P. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dual Coenzyme Specificity of Photosynthetic Glyceraldehyde-3-phosphate
Dehydrogenase Interpreted by the Crystal Structure of A(4) Isoform
Complexed with NAD
Biochemistry, 42, 2003
|
|
6XZF
 
 | |
6XYF
 
 | | Nanobody 22 | | Descriptor: | Nanobody 22, SODIUM ION | | Authors: | Pompidor, G, Zimmermann, S, Loew, C, Schneider, T. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.11097 Å) | | Cite: | Engineered nanobodies with a lanthanide binding motif for crystallographic phasing
To Be Published
|
|
6XYM
 
 | | Nbe-LBM | | Descriptor: | Nbe-LBM, TERBIUM(III) ION | | Authors: | Pompidor, G, Zimmermann, S, Loew, C, Schneider, T. | | Deposit date: | 2020-01-30 | | Release date: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Engineered nanobodies with a lanthanide binding motif for crystallographic phasing
To Be Published
|
|
1SNN
 
 | | 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CALCIUM ION, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Steinbacher, S, Schiffmann, S, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2004-03-11 | | Release date: | 2004-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Metal sites in 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii in complex with the substrate ribulose 5-phosphate.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
8A5Q
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on straight DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin related protein 4 (Arp4), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5P
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on curved DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin related protein 4 (Arp4), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-15 | | Release date: | 2022-12-14 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5A
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of INO80 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-like protein ARP8, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-14 | | Release date: | 2022-12-14 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
8A5D
 
 | | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 | | Descriptor: | Actin, Actin related protein 4 (Arp4), Actin-related protein 8, ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-06-14 | | Release date: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
2YXR
 
 | |
