7WMN
 
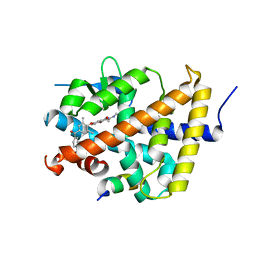 | | A novel chemical derivative(89) of THRB agonist | | Descriptor: | 2-[[7-[2,6-dimethyl-4-(3-methylphenyl)carbonyl-phenoxy]-1-methoxy-4-oxidanyl-isoquinolin-3-yl]carbonylamino]ethanoic acid, Isoform Beta-2 of Thyroid hormone receptor beta, Nuclear receptor coactivator 2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2022-01-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery of a Highly Selective and H435R-Sensitive Thyroid Hormone Receptor beta Agonist.
J.Med.Chem., 65, 2022
|
|
7E7D
 
 | |
7E7B
 
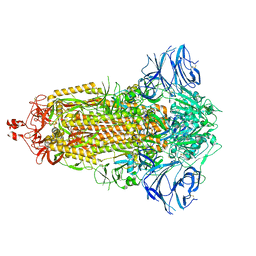
 | | Cryo-EM structure of the SARS-CoV-2 furin site mutant S-Trimer from a subunit vaccine candidate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-hydroxyethyl 2-deoxy-3,5-bis-O-(2-hydroxyethyl)-6-O-(2-{[(9E)-octadec-9-enoyl]oxy}ethyl)-alpha-L-xylo-hexofuranoside, ... | | Authors: | Zheng, S, Ma, J. | | Deposit date: | 2021-02-25 | | Release date: | 2021-03-24 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Cryo-EM structure of S-Trimer, a subunit vaccine candidate for COVID-19.
J.Virol., 95, 2021
|
|
6JXT
 
 | |
6JWL
 
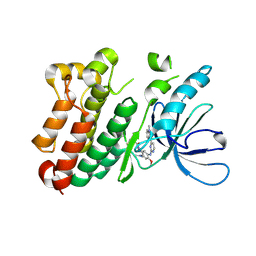
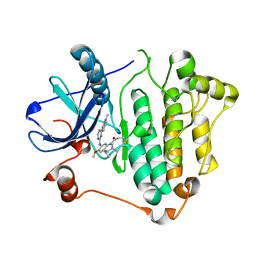 | | Crystal structure of EGFR 696-1022 L858R in complex with AZD9291 | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Yun, C.H, Zhu, S.J, Yan, X.E. | | Deposit date: | 2019-04-21 | | Release date: | 2020-04-22 | | Last modified: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural Basis of AZD9291 Selectivity for EGFR T790M.
J.Med.Chem., 63, 2020
|
|
6JX4
 
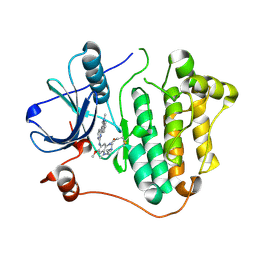 | |
6JX0
 
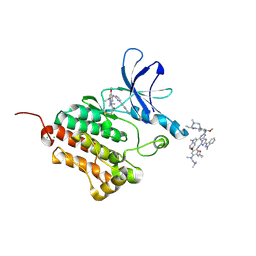 | |
7KEF
 
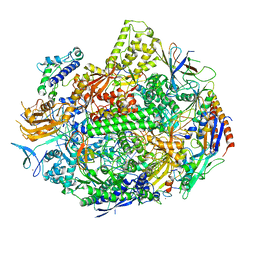 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaM in swing state | | Descriptor: | (1S)-1,4-anhydro-1-(3-methoxynaphthalen-2-yl)-5-O-phosphono-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
7KED
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, W, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
7KEE
 
 | | RNA polymerase II elongation complex with unnatural base dTPT3, rNaMTP bound to E-site | | Descriptor: | (1S)-1,4-anhydro-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-1-(3-methoxynaphthalen-2-yl)-D-ribitol, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2020-10-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Transcriptional processing of an unnatural base pair by eukaryotic RNA polymerase II.
Nat.Chem.Biol., 17, 2021
|
|
8J1V
 
 | |
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DHT
 
 | |
8J1T
 
 | |
7WWH
 
 | |
7YD2
 
 | | SulE_P44R_S209A | | Descriptor: | 2-[(4-chloranyl-6-methoxy-pyrimidin-2-yl)carbamoylsulfamoyl]benzoic acid, 2-[[[[(4-CHLORO-6-METHOXY-2-PYRIMIDINYL)AMINO]CARBONYL]AMINO]SULFONYL]BENZOIC ACID ETHYL ESTER, Alpha/beta fold hydrolase, ... | | Authors: | Liu, B, Ran, T, Wang, W, He, J. | | Deposit date: | 2022-07-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
7Y0L
 
 | | SulE-S209A | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, METHYL 2-[({[(4-METHOXY-6-METHYL-1,3,5-TRIAZIN-2-YL)AMINO]CARBONYL}AMINO)SULFONYL]BENZOATE | | Authors: | Liu, B, Ran, T, He, J, Wang, W. | | Deposit date: | 2022-06-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
