2FTP
 
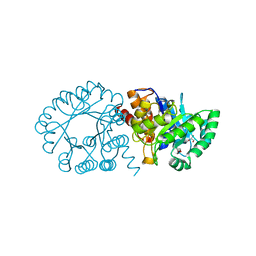 | | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, SODIUM ION, hydroxymethylglutaryl-CoA lyase | | Authors: | Xiao, T, Evdokimova, E, Liu, Y, Kudritska, M, Savchenko, A, Pai, E.F, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-01-24 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of hydroxymethylglutaryl-CoA lyase from Pseudomonas aeruginosa
To be Published
|
|
8I0C
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S0703 | | Descriptor: | 1-[4-[3,5-bis(chloranyl)phenyl]-3-fluoranyl-phenyl]cyclopropane-1-carboxylic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, He, S, Liu, Y, Fang, P, Sun, H. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
3R45
 
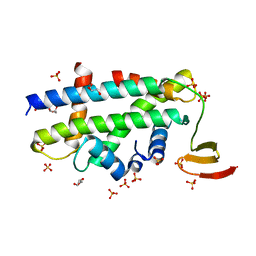 | | Structure of a CENP-A-Histone H4 Heterodimer in complex with chaperone HJURP | | Descriptor: | GLYCEROL, Histone H3-like centromeric protein A, Histone H4, ... | | Authors: | Hu, H, Liu, Y, Wang, M, Fang, J, Huang, H, Yang, N, Li, Y, Wang, J, Yao, X, Shi, Y, Li, G, Xu, R.M. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP
Genes Dev., 25, 2011
|
|
4AZA
 
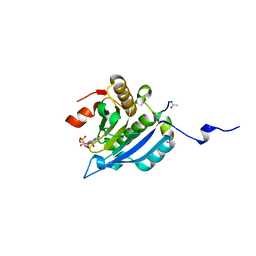 | | Improved eIF4E binding peptides by phage display guided design. | | Descriptor: | EIF4G1_D5S PEPTIDE, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E, [[(2R,3S,4R,5R)-5-(6-AMINO-3-METHYL-4-OXO-5H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHOXY-HYDROXY-PHOSPHORYL] PHOSPHONO HYDROGEN PHOSPHATE | | Authors: | Chew, W.Z, Quah, S.T, Verma, C.S, Liu, Y, Lane, D.P, Brown, C.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Improved Eif4E Binding Peptides by Phage Display Guided Design: Plasticity of Interacting Surfaces Yield Collective Effects.
Plos One, 7, 2012
|
|
1Y4E
 
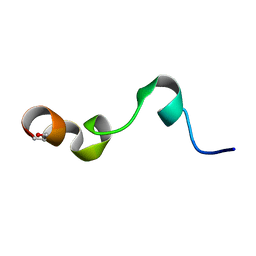 | | NMR structure of transmembrane segment IV of the NHE1 isoform of the Na+/H+ exchanger | | Descriptor: | Sodium/hydrogen exchanger 1 | | Authors: | Slepkov, E.R, Rainey, J.K, Li, X, Liu, Y, Lindhout, D.A, Sykes, B.D, Fliegel, L. | | Deposit date: | 2004-11-30 | | Release date: | 2005-02-01 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of transmembrane segment IV of the NHE1 isoform of the Na+/H+ exchanger.
J.Biol.Chem., 280, 2005
|
|
4CUS
 
 | | Crystal structure of human BAZ2B in complex with fragment-4 N09496 | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2B, quinolin-4-ol | | Authors: | Bradley, A.R, Liu, Y, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, von Delft, F. | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Crystal Structure of Human Baz2B in Complex with Fragment-4 N09496
To be Published
|
|
4CUT
 
 | | Crystal structure of human BAZ2B in complex with fragment-5 N09428 | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2B, N-(4-HYDROXYPHENYL)ACETAMIDE (TYLENOL) | | Authors: | Bradley, A.R, Liu, Y, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, von Delft, F. | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Crystal Structure of Human Baz2B in Complex with Fragment-5 N09428
To be Published
|
|
1Y4U
 
 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | Chaperone protein htpG | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
4D0Q
 
 | | Hyaluronan Binding Module of the Streptococcal Pneumoniae Hyaluronate Lyase | | Descriptor: | 1,2-ETHANEDIOL, HYALURONATE LYASE | | Authors: | Suits, M.D.L, Pluvinage, B, Law, A, Liu, Y, Palma, A.S, Chai, W, Feizi, T, Boraston, A.B. | | Deposit date: | 2014-04-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Conformational Analysis of the Streptococcus Pneumoniae Hyaluronate Lyase and Characterization of its Hyaluronan-Specific Carbohydrate-Binding Module.
J.Biol.Chem., 289, 2014
|
|
1Y4S
 
 | | Conformation rearrangement of heat shock protein 90 upon ADP binding | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein htpG, MAGNESIUM ION | | Authors: | Huai, Q, Wang, H, Liu, Y, Kim, H, Toft, D, Ke, H. | | Deposit date: | 2004-12-01 | | Release date: | 2005-04-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the N-terminal and middle domains of E. coli Hsp90 and conformation changes upon ADP binding.
Structure, 13, 2005
|
|
4CUP
 
 | | Crystal structure of human BAZ2B in complex with fragment-1 N09421 | | Descriptor: | 4-Fluorobenzamidoxime, BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2B, METHANOL | | Authors: | Bradley, A.R, Liu, Y, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, von Delft, F. | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of Human Baz2B in Complex with Fragment-1 N09421
To be Published
|
|
6M4P
 
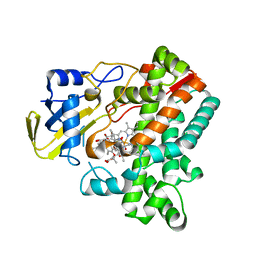 | | Cytochrome P450 monooxygenase StvP2 substrate-bound structure | | Descriptor: | 6-methoxy-streptovaricin C, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | Deposit date: | 2020-03-08 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
4ZOU
 
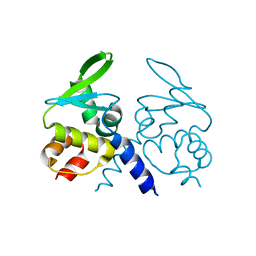 | |
6ND7
 
 | | The crystal structure of TerB co-crystallized with polyporic acid | | Descriptor: | 2~3~,2~6~-dihydroxy[1~1~,2~1~:2~4~,3~1~-terphenyl]-2~2~,2~5~-dione, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Clinger, J.A, Elshahawi, S.I, Zhang, Y, Hall, R.P, Liu, Y, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2018-12-13 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure and Function of Terfestatin Biosynthesis Enzymes TerB and TerC
To Be Published
|
|
6NN3
 
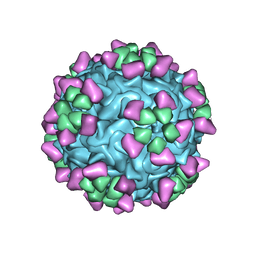 | | Structure of parvovirus B19 decorated with Fab molecules from a human antibody | | Descriptor: | Fab monoclonal antibody 860-55D, heavy chain, light chain, ... | | Authors: | Rossmann, M.G, Sun, Y, Klose, T, Liu, Y. | | Deposit date: | 2019-01-14 | | Release date: | 2019-02-13 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structure of Parvovirus B19 Decorated by Fabs from a Human Antibody.
J. Virol., 93, 2019
|
|
4ZUZ
 
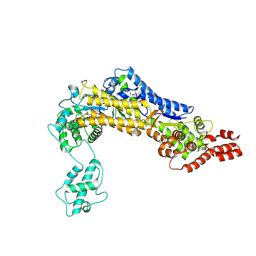 | | SidC 1-871 | | Descriptor: | SidC | | Authors: | Luo, X, Wasilko, D.J, Liu, Y, Sun, J, Wu, X, Luo, Z.-Q, Mao, Y. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of the Legionella Virulence Factor, SidC Reveals a Unique PI(4)P-Specific Binding Domain Essential for Its Targeting to the Bacterial Phagosome.
Plos Pathog., 11, 2015
|
|
9KMH
 
 | | The Composite Cryo-EM Structure of the Portal Vertex of Bacteriophage FCWL1 | | Descriptor: | Adaptor protein, Connector protein, Decoration protein, ... | | Authors: | Cai, C, Wang, Y, Liu, Y, Shao, Q, Wang, A, Fang, Q. | | Deposit date: | 2024-11-16 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of a T1-like siphophage reveal capsid stabilization mechanisms and high structural similarities with a myophage.
Structure, 33, 2025
|
|
8K58
 
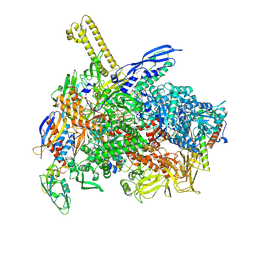 | | The cryo-EM map of close TIEA-TEC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | TIEA inhibits Sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
8K5A
 
 | | The cryo-EM map of open TIEA-TEC complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | TIEA inhibits sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
9JLF
 
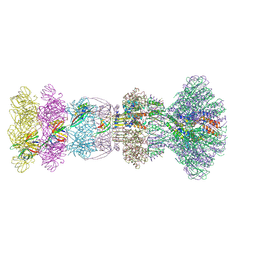 | | Cryo-EM Structure of Bacteriophage FCWL1 head-to-tail interface | | Descriptor: | Adaptor protein, Connector protein, Portal protein, ... | | Authors: | Cai, C, Wang, Y, Liu, Y, Shao, Q, Wang, A, Fang, Q. | | Deposit date: | 2024-09-18 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of a T1-like siphophage reveal capsid stabilization mechanisms and high structural similarities with a myophage.
Structure, 33, 2025
|
|
9KMG
 
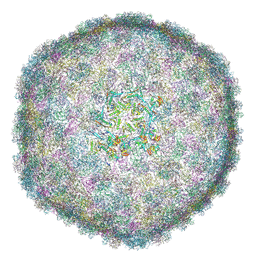 | | Cryo-EM Structure of Bacteriophage FCWL1 Capsid | | Descriptor: | Decoration protein, Major capsid protein | | Authors: | Cai, C, Wang, Y, Liu, Y, Shao, Q, Wang, A, Fang, Q. | | Deposit date: | 2024-11-16 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of a T1-like siphophage reveal capsid stabilization mechanisms and high structural similarities with a myophage.
Structure, 33, 2025
|
|
8K59
 
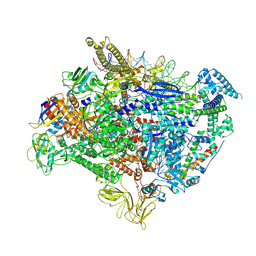 | | The cryo-EM map of TIC-TIEA complex | | Descriptor: | 15 kDa RNA polymerase-binding protein, DNA (61-MER), DNA (63-MER), ... | | Authors: | Zhang, K.N, Liu, Y, Chen, M, Wang, Y, Lin, W, Li, M, Zhang, X, Gao, Y, Gong, Q, Chen, H, Steve, M, Li, S, Zhang, K, Liu, B. | | Deposit date: | 2023-07-21 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | TIEA inhibits sigma70-dependent transcriptions, accelerates elongation speed and elevates transcription error
To Be Published
|
|
5C6H
 
 | | Mcl-1 complexed with Mule | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mule BH3 peptide from E3 ubiquitin-protein ligase HUWE1 | | Authors: | Song, T, Wang, Z, Ji, F, Chai, G, Liu, Y, Li, X, Li, Z, Fan, Y, Zhang, Z. | | Deposit date: | 2015-06-23 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Mcl-1 complexed with Mule at 2.05 Angstroms resolution
To Be Published
|
|
1Q67
 
 | | Crystal structure of Dcp1p | | Descriptor: | Decapping protein involved in mRNA degradation-Dcp1p | | Authors: | She, M, Decker, C.J, Liu, Y, Chen, N, Parker, R, Song, H. | | Deposit date: | 2003-08-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Dcp1p and its functional implications in mRNA decapping
Nat.Struct.Mol.Biol., 11, 2004
|
|
1T1J
 
 | | Crystal structure of genomics APC5043 | | Descriptor: | hypothetical protein | | Authors: | Dong, A, Xu, X, Liu, Y, Zhang, R, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-04-16 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Conserved Hypothetical Protein PA1492 from Pseudomonas aeruginosa
TO BE PUBLISHED
|
|
