7PSE
 
 | | Crystal Structure of a Class D Carbapenemase_K73ALY Complexed with Oxacillin | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, 1-BUTANOL, Beta-lactamase, ... | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-09-23 | | Release date: | 2022-10-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
8Q1K
 
 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Roske, Y, Daumke, O, Damme, M. | | Deposit date: | 2023-07-31 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
3U0M
 
 | |
8Q1X
 
 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Roske, Y, Daumke, O, Damme, M. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
7Q14
 
 | |
7W6P
 
 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a G protein biased agonist | | Descriptor: | Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
7W7E
 
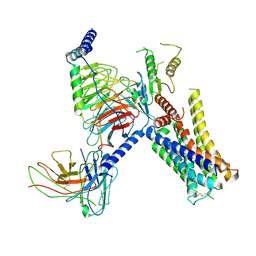 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a biased agonist | | Descriptor: | 5-(3-bicyclo[4.2.0]octa-1,3,5-trienyl)-1,2,3,6-tetrahydropyridine, Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
3U0N
 
 | |
3U0K
 
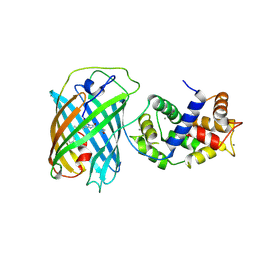 | |
7PUX
 
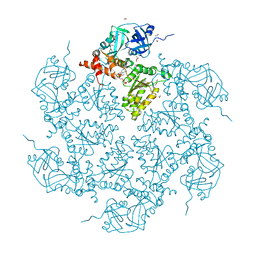 | | Structure of p97 N-D1(L198W) in complex with Fragment TROLL2 | | Descriptor: | (1S)-2-amino-1-(4-bromophenyl)ethan-1-ol, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bothe, S, Schindelin, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment screening using biolayer interferometry reveals ligands targeting the SHP-motif binding site of the AAA+ ATPase p97
Commun Chem, 5, 2022
|
|
3HBR
 
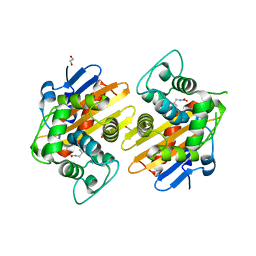 | | Crystal structure of OXA-48 beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, OXA-48 | | Authors: | Calderone, V, Mangani, S, Benvenuti, M, Rossolini, G.M, Docquier, J.D. | | Deposit date: | 2009-05-05 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the OXA-48 beta-lactamase reveals mechanistic diversity among class D carbapenemases.
Chem.Biol., 16, 2009
|
|
5NCF
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-4]-OH | | Descriptor: | (1~{S},4~{S},7~{R},10~{R})-14-[(3~{S},6~{R},8~{a}~{S})-1'-[(2~{S})-2-acetamido-3-(2-chlorophenyl)propanoyl]-5-oxidanylidene-spiro[1,2,3,8~{a}-tetrahydroindolizine-6,2'-pyrrolidine]-3-yl]carbonyl-2-oxidanylidene-3,14-diazatricyclo[8.4.0.0^{3,7}]tetradec-8-ene-4-carboxylic acid, GLYCEROL, NITRATE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-03-04 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5N9P
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PP-[ProM-1]-NH2 | | Descriptor: | Ac-[2-Cl-F]-PP-[ProM-1]-NH2, CHLORIDE ION, Protein enabled homolog | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-26 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5HFO
 
 | | CRYSTAL STRUCTURE OF OXA-232 BETA-LACTAMASE | | Descriptor: | Beta-lactamase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Retailleau, P, Oueslati, S, Cisse, C, Nordmann, P, Naas, T, Iorga, B. | | Deposit date: | 2016-01-07 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Role of Arginine 214 in the Substrate Specificity of OXA-48.
Antimicrob.Agents Chemother., 64, 2020
|
|
5N9C
 
 | | ENAH EVH1 in complex with Ac-[2-Cl-F]-PP-[ProM-1]-OH | | Descriptor: | Ac-[2-Cl-F]-PP-[ProM-1]-OH, GLYCEROL, NITRATE ION, ... | | Authors: | Barone, M, Roske, Y. | | Deposit date: | 2017-02-24 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Designed nanomolar small-molecule inhibitors of Ena/VASP EVH1 interaction impair invasion and extravasation of breast cancer cells.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6SUR
 
 | | The Rab33B-Atg16L1 crystal structure | | Descriptor: | Autophagy-related protein 16-1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Metje-Sprink, J, Kuehnel, K. | | Deposit date: | 2019-09-16 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.467 Å) | | Cite: | Crystal structure of the Rab33B/Atg16L1 effector complex.
Sci Rep, 10, 2020
|
|
8AAK
 
 | | Crystal structure of the PDZ tandem of syntenin in complex with compound 29 | | Descriptor: | (2~{S})-2-[[(2~{S})-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)-3-phenyl-propanoyl]amino]propanoic acid, GLYCEROL, Syntenin-1 | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Discovery of a PDZ Domain Inhibitor Targeting the Syndecan/Syntenin Protein-Protein Interaction: A Semi-Automated "Hit Identification-to-Optimization" Approach.
J.Med.Chem., 66, 2023
|
|
8AAP
 
 | | Crystal structure of the PDZ tandem of syntenin in complex with compound SYNTi | | Descriptor: | (2S)-2-[[(2S)-2-(6-bromanyl-3-oxidanylidene-1H-isoindol-2-yl)-3-[4-(5-ethanoyl-2-fluoranyl-phenyl)phenyl]propanoyl]amino]propanoic acid, Syntenin-1 | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Discovery of a PDZ Domain Inhibitor Targeting the Syndecan/Syntenin Protein-Protein Interaction: A Semi-Automated "Hit Identification-to-Optimization" Approach.
J.Med.Chem., 66, 2023
|
|
8AAO
 
 | | Crystal structure of the PDZ tandem of syntenin in complex with compound 95 | | Descriptor: | (2~{S})-2-[[(2~{S})-3-[4-(5-ethanoyl-2-fluoranyl-phenyl)phenyl]-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)propanoyl]amino]propanoic acid, Syntenin-1 | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Discovery of a PDZ Domain Inhibitor Targeting the Syndecan/Syntenin Protein-Protein Interaction: A Semi-Automated "Hit Identification-to-Optimization" Approach.
J.Med.Chem., 66, 2023
|
|
8AAI
 
 | | Crystal structure of the PDZ tandem of syntenin in complex with fragment E5 | | Descriptor: | (2~{S})-2-[[(2~{S})-3-methyl-2-(3-oxidanylidene-1~{H}-isoindol-2-yl)butanoyl]amino]propanoic acid, Syntenin-1 | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of a PDZ Domain Inhibitor Targeting the Syndecan/Syntenin Protein-Protein Interaction: A Semi-Automated "Hit Identification-to-Optimization" Approach.
J.Med.Chem., 66, 2023
|
|
5WHR
 
 | | Discovery of a novel and selective IDO-1 inhibitor PF-06840003 and its characterization as a potential clinical candidate. | | Descriptor: | (3R)-3-(5-fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Greasley, S.E, Kaiser, S.E, Feng, J.L, Stewart, A. | | Deposit date: | 2017-07-18 | | Release date: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of a Novel and Selective Indoleamine 2,3-Dioxygenase (IDO-1) Inhibitor 3-(5-Fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione (EOS200271/PF-06840003) and Its Characterization as a Potential Clinical Candidate.
J. Med. Chem., 60, 2017
|
|
6TVO
 
 | | Human CRM1-RanGTP in complex with Leptomycin B | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shaikhqasem, A, Ficner, R. | | Deposit date: | 2020-01-10 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Characterization of Inhibition Reveals Distinctive Properties for Human andSaccharomyces cerevisiaeCRM1.
J.Med.Chem., 63, 2020
|
|
2VSF
 
 | | Structure of XPD from Thermoplasma acidophilum | | Descriptor: | CALCIUM ION, DNA REPAIR HELICASE RAD3 RELATED PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Kuper, J, Wolski, S.C, Truglio, J.J, Kisker, C. | | Deposit date: | 2008-04-23 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of the Fes Cluster-Containing Nucleotide Excision Repair Helicase Xpd.
Plos Biol., 6, 2008
|
|
5OQ4
 
 | | PQR309 - a Potent, Brain-Penetrant, Orally Bioavailable, pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology | | Descriptor: | 5-(4,6-dimorpholin-4-yl-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Williams, R.L, Zhang, X. | | Deposit date: | 2017-08-10 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-(4,6-Dimorpholino-1,3,5-triazin-2-yl)-4-(trifluoromethyl)pyridin-2-amine (PQR309), a Potent, Brain-Penetrant, Orally Bioavailable, Pan-Class I PI3K/mTOR Inhibitor as Clinical Candidate in Oncology.
J. Med. Chem., 60, 2017
|
|
8PSV
 
 | | 2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod | | Descriptor: | Type-1 fimbrial protein, A chain | | Authors: | Hospenthal, M, Zyla, D, Glockshuber, R, Waksman, G. | | Deposit date: | 2023-07-13 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili.
Nat Commun, 15, 2024
|
|
