7V6U
 
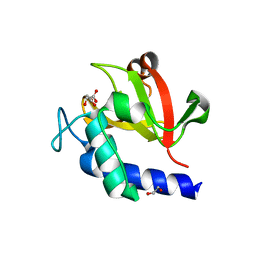 | | Crystal structure of bacterial peptidase | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.144 Å) | | Cite: | The crystal structure of bacterial DD-endopeptidase
To be published
|
|
7V6T
 
 | | Crystal structure of bacterial peptidase | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Crystal structure of bacterial DD-endopeptidase
To be published
|
|
7V6S
 
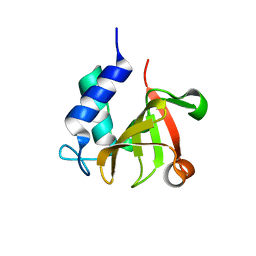 | | Crystal structure of bacterial peptidase | | Descriptor: | Murein DD-endopeptidase MepS/Murein LD-carboxypeptidase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.883 Å) | | Cite: | The crystal structure fre
To be published
|
|
7CAZ
 
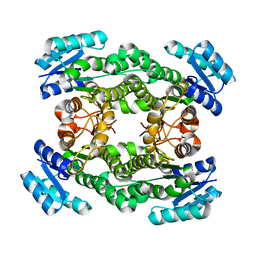 | | Crystal structure of bacterial reductase | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, GLYCEROL | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of bacterial reductase
To be published
|
|
7CAW
 
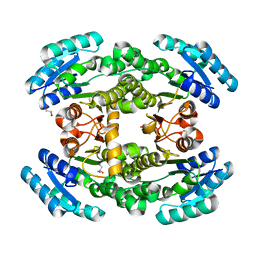 | | Crystal structure of bacterial reductase | | Descriptor: | 3-oxoacyl-ACP reductase FabG, GLYCEROL | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | Crystal structure of bacterial reductase
To be published
|
|
7CAX
 
 | | Crystal structure of bacterial reductase | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-ACP reductase FabG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Crystal structure of bacterial reductase
To be published
|
|
7F27
 
 | | Crystal structure of polyketide ketosynthase | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) synthase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis.
Sci Rep, 11, 2021
|
|
5WHM
 
 | | Crystal Structure of IclR Family Transcriptional Regulator from Brucella abortus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Kim, Y, Wu, R, Tesar, C, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-07-17 | | Release date: | 2017-08-23 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular control of gene expression byBrucellaBaaR, an IclR-type transcriptional repressor.
J. Biol. Chem., 293, 2018
|
|
5WEJ
 
 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with a dipeptidyl oxazolidinone-based inhibitor | | Descriptor: | (2S)-2-{(5S)-5-[(3-chlorophenyl)methyl]-2-oxo-1,3-oxazolidin-3-yl}-4-methyl-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}pentanamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Kankanamalage, A.C.G, Rathnayake, A.D, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2017-07-10 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-guided design, synthesis and evaluation of oxazolidinone-based inhibitors of norovirus 3CL protease.
Eur J Med Chem, 143, 2017
|
|
2E2E
 
 | | TPR domain of NrfG mediates the complex formation between heme lyase and formate-dependent nitrite reductase in Escherichia Coli O157:H7 | | Descriptor: | BETA-MERCAPTOETHANOL, Formate-dependent nitrite reductase complex nrfG subunit, IMIDAZOLE | | Authors: | Han, D, Kim, K, Oh, J, Park, J, Kim, Y. | | Deposit date: | 2006-11-11 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | TPR domain of NrfG mediates complex formation between heme lyase and formate-dependent nitrite reductase in Escherichia coli O157:H7.
Proteins, 70, 2008
|
|
1U14
 
 | | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom | | Descriptor: | Hypothetical UPF0244 protein yjjX, PHOSPHATE ION | | Authors: | Qiu, Y, Kim, Y, Cuff, M, Collart, F, Joachimiak, A, Kossiakoff, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The crystal structure of hypothetical UPF0244 protein yjjX at resolution 1.68 Angstrom
To be Published
|
|
6E3S
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Antagonist PT2385 | | Descriptor: | 3-{[(1S)-2,2-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
6E3T
 
 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Antagonist T1001 | | Descriptor: | (6S)-6-(4-bromophenyl)-2,3,5,6-tetrahydroimidazo[2,1-b][1,3]thiazole, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
6E3U
 
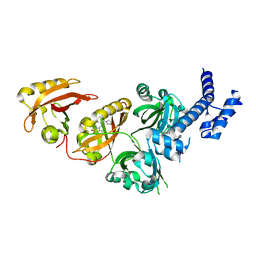 | | Crystal Structure of the Heterodimeric HIF-2 Complex with Agonist M1001 | | Descriptor: | 3-{[2-(pyrrolidin-1-yl)phenyl]amino}-1H-1lambda~6~,2-benzothiazole-1,1-dione, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Wu, D, Su, X, Lu, J, Li, S, Hood, B, Vasile, S, Potluri, N, Diao, X, Kim, Y, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 2018-07-15 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bidirectional modulation of HIF-2 activity through chemical ligands.
Nat. Chem. Biol., 15, 2019
|
|
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
3TBF
 
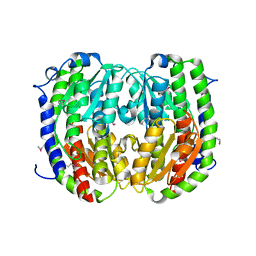 | | C-terminal domain of glucosamine-fructose-6-phosphate aminotransferase from Francisella tularensis. | | Descriptor: | Glucosamine--fructose-6-phosphate aminotransferase [isomerizing] | | Authors: | Osipiuk, J, Zhou, M, Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-08-05 | | Release date: | 2011-08-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | C-terminal domain of glucosamine-fructose-6-phosphate aminotransferase from Francisella tularensis.
To be Published
|
|
3FH3
 
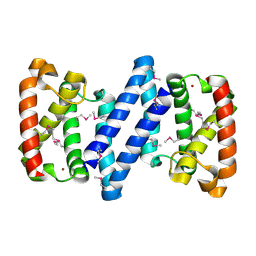 | | Crystal structure of a putative ECF-type sigma factor negative effector from Bacillus anthracis str. Sterne | | Descriptor: | NICKEL (II) ION, putative ECF-type sigma factor negative effector | | Authors: | Nocek, B, Kim, Y, Joachimiak, G, Du, J, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2009-01-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of a putative ECF-type sigma factor negative effector from Bacillus anthracis str. Sterne
To be Published
|
|
3FFY
 
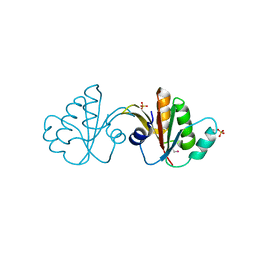 | | Putative tetrapyrrole (corrin/porphyrin) methyltransferase from Bacteroides fragilis. | | Descriptor: | Putative tetrapyrrole (Corrin/porphyrin) methylase, SULFATE ION | | Authors: | Osipiuk, J, Volkart, L, Cobb, G, Kim, Y, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of putative tetrapyrrole (corrin/porphyrin) methyltransferase from Bacteroides fragilis.
To be Published
|
|
1U13
 
 | |
6LHU
 
 | | High resolution structure of FANCA C-terminal domain (CTD) | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6LHW
 
 | | Structure of N-terminal and C-terminal domains of FANCA | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.84 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
4ZQD
 
 | | Crystal Structure of the Heterodimeric HIF-2a:ARNT Complex with the Benzoxadiazole Antagonist 0X3 | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(3-chloro-5-fluorophenyl)-4-nitro-2,1,3-benzoxadiazol-5-amine | | Authors: | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | Deposit date: | 2015-05-09 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
4ZPR
 
 | | Crystal Structure of the Heterodimeric HIF-1a:ARNT Complex with HRE DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*CP*AP*CP*GP*AP*CP*CP*CP*GP*CP*AP*CP*GP*TP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*GP*CP*GP*TP*AP*CP*GP*TP*GP*CP*GP*GP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | Deposit date: | 2015-05-08 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.902 Å) | | Cite: | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
4ZPK
 
 | | Crystal Structure of the Heterodimeric HIF-2a:ARNT Complex with HRE DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*CP*AP*CP*GP*AP*CP*CP*CP*GP*CP*AP*CP*GP*TP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*GP*CP*GP*TP*AP*CP*GP*TP*GP*CP*GP*GP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Wu, D, Potluri, N, Lu, J, Kim, Y, Rastinejad, F. | | Deposit date: | 2015-05-08 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural integration in hypoxia-inducible factors.
Nature, 524, 2015
|
|
