7WOG
 
 | | SARS-CoV-2 Omicron S monomer complexed with 553-49 | | Descriptor: | 553-49 VH, 553-49 VL, Spike protein S1 | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WO4
 
 | | SARS-CoV-2 Spike in complex with IgG 553-15 (S-553-15 dimer trimer ) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb15 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.47 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WO7
 
 | | Locally refined region of SARS-CoV-2 Spike in complex with IgG 553-15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, mAb15 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WZ2
 
 | | SARS-CoV-2 (D614G) Spike trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-02-16 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WO5
 
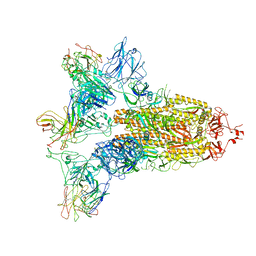 | | SARS-CoV-2 Spike in complex with IgG 553-15 (S-553-15 trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb15 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-20 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WOC
 
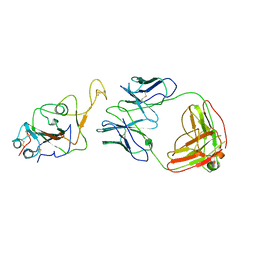 | | Locally refined region of SARS-CoV-2 Spike in complex with IgG 553-60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, mAb60 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WOA
 
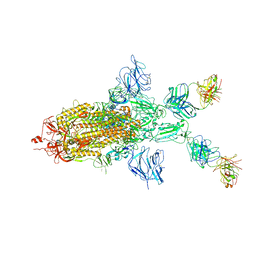 | | SARS-CoV-2 Spike in complex with IgG 553-60 (1-up trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, mAb60 VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-01-21 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
7WZ1
 
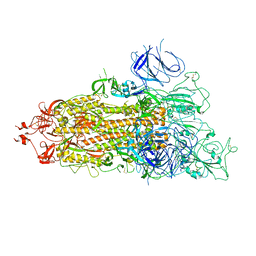 | | SARS-CoV-2 Omicron Spike trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2022-02-16 | | Release date: | 2022-07-27 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Study of SARS-CoV-2 Antibodies Identifies a Broad-Spectrum Antibody That Neutralizes the Omicron Variant by Disassembling the Spike Trimer.
J.Virol., 96, 2022
|
|
8D95
 
 | |
7YV4
 
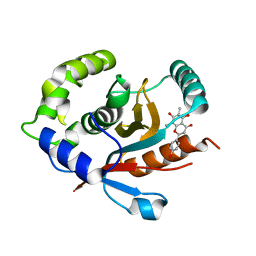 | | Crystal structure of human UCHL3 in complex with Farrerol | | Descriptor: | (2~{S})-2-(4-hydroxyphenyl)-6,8-dimethyl-5,7-bis(oxidanyl)-2,3-dihydrochromen-4-one, Ubiquitin carboxyl-terminal hydrolase isozyme L3 | | Authors: | Mao, Z.Y, Xu, X.J, Zhang, W.T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Farrerol directly activates the deubiqutinase UCHL3 to promote DNA repair and reprogramming when mediated by somatic cell nuclear transfer.
Nat Commun, 14, 2023
|
|
9J4X
 
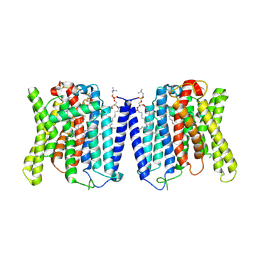 | | CryoEM structure of human XPR1 in apo state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Solute carrier family 53 member 1 | | Authors: | Zhang, W.H, Chen, Y.K, Guan, Z.Y, Liu, Z. | | Deposit date: | 2024-08-10 | | Release date: | 2024-12-04 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the mechanism of phosphate recognition and transport by XPR1.
Nat Commun, 16, 2025
|
|
6OCW
 
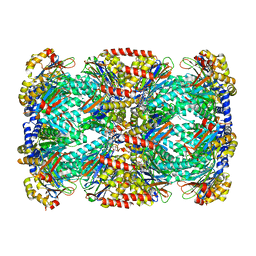 | | Crystal Structure of Mycobacterium tuberculosis Proteasome in Complex with Phenylimidazole-based Inhibitor A85 | | Descriptor: | CITRIC ACID, DIMETHYLFORMAMIDE, N-{(2S)-1-({(2S)-1-[(2,4-difluorobenzyl)amino]-1-oxopropan-2-yl}amino)-4-[(2S)-2-methylpiperidin-1-yl]-1,4-dioxobutan-2-yl}-5-methyl-1,2-oxazole-3-carboxamide (non-preferred name), ... | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2019-03-25 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective Phenylimidazole-Based Inhibitors of theMycobacterium tuberculosisProteasome.
J.Med.Chem., 62, 2019
|
|
9J52
 
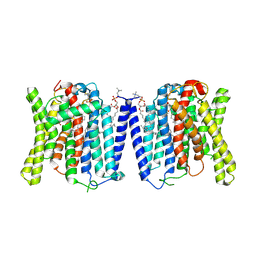 | | CryoEM structure of human XPR1 in complex with phosphate in state B | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PHOSPHATE ION, Solute carrier family 53 member 1 | | Authors: | Zhang, W.H, Chen, Y.K, Guan, Z.Y, Liu, Z. | | Deposit date: | 2024-08-11 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanism of phosphate recognition and transport by XPR1.
Nat Commun, 16, 2025
|
|
9J53
 
 | | CryoEM structure of human XPR1 in complex with phosphate in state C | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PHOSPHATE ION, Solute carrier family 53 member 1 | | Authors: | Zhang, W.H, Chen, Y.K, Guan, Z.Y, Liu, Z. | | Deposit date: | 2024-08-11 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the mechanism of phosphate recognition and transport by XPR1.
Nat Commun, 16, 2025
|
|
9J51
 
 | | CryoEM structure of human XPR1 in complex with phosphate in state A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, PHOSPHATE ION, Solute carrier family 53 member 1 | | Authors: | Zhang, W.H, Chen, Y.K, Guan, Z.Y, Liu, Z. | | Deposit date: | 2024-08-11 | | Release date: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the mechanism of phosphate recognition and transport by XPR1.
Nat Commun, 16, 2025
|
|
6ODE
 
 | |
5V69
 
 | |
5V5G
 
 | |
8SRO
 
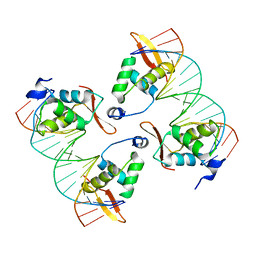 | | FoxP3 tetramer on TTTG repeats | | Descriptor: | DNA 72-mer, Forkhead box protein P3 | | Authors: | Leng, F, Hur, S. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | FOXP3 recognizes microsatellites and bridges DNA through multimerization.
Nature, 624, 2023
|
|
8SRP
 
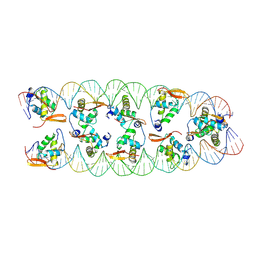 | |
5V5I
 
 | |
5V5H
 
 | |
8I71
 
 | | Hepatitis B virus core protein Y132A mutant in complex with Linvencorvir (RG7907), a Hepatitis B Virus (HBV) Core Protein Allosteric Modulator (CpAM) | | Descriptor: | 3-[(8~{a}~{S})-7-[[5-ethoxycarbonyl-4-(3-fluoranyl-2-methyl-phenyl)-2-(1,3-thiazol-2-yl)-1,4-dihydropyrimidin-6-yl]methyl]-3-oxidanylidene-5,6,8,8~{a}-tetrahydro-1~{H}-imidazo[1,5-a]pyrazin-2-yl]-2,2-dimethyl-propanoic acid, CHLORIDE ION, Capsid protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Linvencorvir (RG7907), a Hepatitis B Virus Core Protein Allosteric Modulator, for the Treatment of Chronic HBV Infection.
J.Med.Chem., 66, 2023
|
|
6OCZ
 
 | | Crystal Structure of Mycobacterium tuberculosis Proteasome in Complex with Phenylimidazole-based Inhibitor A86 | | Descriptor: | CITRIC ACID, DIMETHYLFORMAMIDE, N-{(2S)-1-({(2S)-1-[(2,4-difluorobenzyl)amino]-1-oxopropan-2-yl}amino)-1,4-dioxo-4-[(2R)-2-phenylpyrrolidin-1-yl]butan-2-yl}-5-methyl-1,2-oxazole-3-carboxamide (non-preferred name), ... | | Authors: | Hsu, H.C, Li, H. | | Deposit date: | 2019-03-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Selective Phenylimidazole-Based Inhibitors of theMycobacterium tuberculosisProteasome.
J.Med.Chem., 62, 2019
|
|
5V6A
 
 | |
