7O0Z
 
 | | ABC transporter NosFY, nucleotide-free in GDN | | Descriptor: | Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter permease protein NosY | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O13
 
 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
5DUT
 
 | | Influenza A virus H5 hemagglutinin globular head | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Zuo, T, Sun, J, Wang, G, Zhou, P, Wang, X, Zhang, L. | | Deposit date: | 2015-09-20 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection
Nat Commun, 6, 2015
|
|
6G3O
 
 | | Crystal structure of human HDAC2 in complex with (R)-6-[3,4-Dioxo-2-(4-trifluoromethoxy-phenylamino)-cyclobut-1-enylamino]-heptanoic acid hydroxyamide | | Descriptor: | (6~{R})-6-[[3,4-bis(oxidanylidene)-2-[[4-(trifluoromethyloxy)phenyl]amino]cyclobuten-1-yl]amino]-~{N}-oxidanyl-heptanamide, CALCIUM ION, Histone deacetylase 2, ... | | Authors: | Isabet, T, Aurelly, M, Chantalat, L, Thoreau, E. | | Deposit date: | 2018-03-26 | | Release date: | 2018-06-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Squaramides as novel class I and IIB histone deacetylase inhibitors for topical treatment of cutaneous t-cell lymphoma.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
4GPZ
 
 | |
7CDJ
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1A3 with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1A3 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Ge, J, Wang, R. | | Deposit date: | 2020-06-19 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.396 Å) | | Cite: | Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry.
Nat Commun, 12, 2021
|
|
4GWK
 
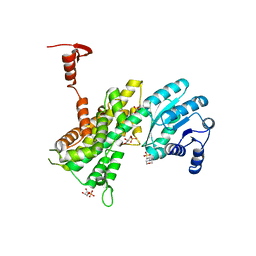 | | Crystal structure of 6-phosphogluconate dehydrogenase complexed with 3-phosphoglyceric acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-PHOSPHOGLYCERIC ACID, 6-phosphogluconate dehydrogenase, ... | | Authors: | He, C, Zhou, L, Zhang, L. | | Deposit date: | 2012-09-03 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth.
Cancer Cell, 22, 2012
|
|
7XT4
 
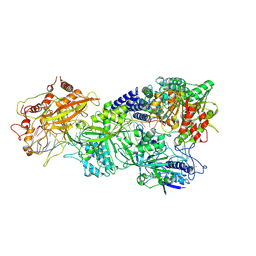 | | Structure of Craspase-NTR | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-16 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7XSO
 
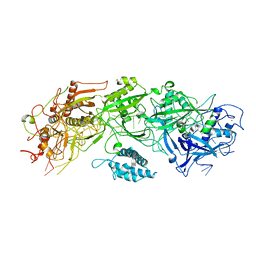 | | Structure of the type III-E CRISPR-Cas effector gRAMP | | Descriptor: | RAMP superfamily protein, RNA (35-MER), ZINC ION | | Authors: | Feng, Y, Zhang, L. | | Deposit date: | 2022-05-15 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
6MVD
 
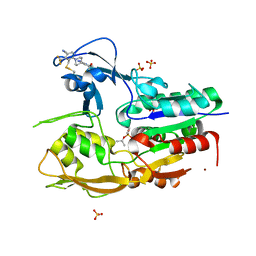 | | Crystal structure of Lecithin:cholesterol acyltransferase (LCAT) in complex with isopropyl dodec-11-enylfluorophosphonate (IDFP) and a small molecule activator | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-{4-[(4R)-4-hydroxy-6-oxo-4-(trifluoromethyl)-4,5,6,7-tetrahydro-2H-pyrazolo[3,4-b]pyridin-3-yl]piperidin-1-yl}-4-(trifluoromethyl)pyridine-3-carbonitrile, NICKEL (II) ION, ... | | Authors: | Manthei, K.A, Chang, L, Tesmer, J.J.G. | | Deposit date: | 2018-10-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular basis for activation of lecithin:cholesterol acyltransferase by a compound that increases HDL cholesterol.
Elife, 7, 2018
|
|
7YXU
 
 | | Crystal structure of agonistic antibody 1618 fab domain bound to human 4-1BB. | | Descriptor: | MANGANESE (II) ION, Tumor necrosis factor receptor superfamily member 9, heavy chain of Fab, ... | | Authors: | Hakansson, M, Rose, N, Petersson, J, Enell Smith, K, Thorolfsson, M, von Schantz, L. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Bispecific Tumor Antigen-Conditional 4-1BB x 5T4 Agonist, ALG.APV-527, Mediates Strong T-Cell Activation and Potent Antitumor Activity in Preclinical Studies.
Mol.Cancer Ther., 22, 2023
|
|
5UP9
 
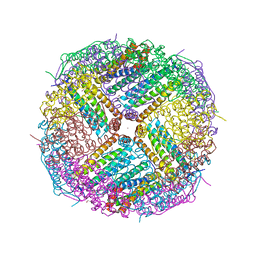 | | Crystal Structure of Zn-bound Human Heavy-Chain ferritin variant 122H-delta C-star with para-xylenedihydroxamate | | Descriptor: | 2,2'-(1,4-phenylene)bis(N-hydroxyacetamide), DI(HYDROXYETHYL)ETHER, Ferritin heavy chain, ... | | Authors: | Bailey, J.B, Zhang, L, Chiong, J.A, Ahn, S, Tezcan, F.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthetic Modularity of Protein-Metal-Organic Frameworks.
J. Am. Chem. Soc., 139, 2017
|
|
5UP8
 
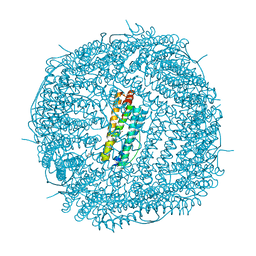 | | Crystal Structure of the Zn-bound Human Heavy-Chain variant 122H-delta C-star with para-benzenedihydroxamate | | Descriptor: | Ferritin heavy chain, N,N'-dihydroxybenzene-1,4-dicarboxamide, SODIUM ION, ... | | Authors: | Bailey, J.B, Zhang, L, Chiong, J.C, Ahn, S, Tezcan, F.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Synthetic Modularity of Protein-Metal-Organic Frameworks.
J. Am. Chem. Soc., 139, 2017
|
|
5VTD
 
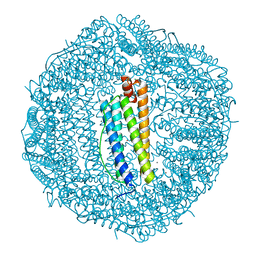 | | Crystal Structure of the Co-bound Human Heavy-Chain Ferritin variant 122H-delta C-star | | Descriptor: | CALCIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Bailey, J.B, Zhang, L, Chiong, J.A, Tezcan, F.A. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Synthetic Modularity of Protein-Metal-Organic Frameworks.
J. Am. Chem. Soc., 139, 2017
|
|
7D5Z
 
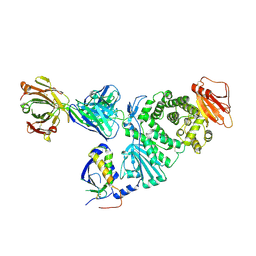 | | Crystal structure of EBV gH/gL bound with neutralizing antibody 1D8 | | Descriptor: | Envelope glycoprotein H, Envelope glycoprotein L, heavy chain of 1D8, ... | | Authors: | Zhu, Q, Shan, S, Yu, J, Wang, X, Zhang, L, Zeng, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | A Neutralizing Antibody Targeting a New Site of Vulnerability on Epstein-Barr Virus gH/gL Protects against Dual-Tropic Infection
To Be Published
|
|
5UP7
 
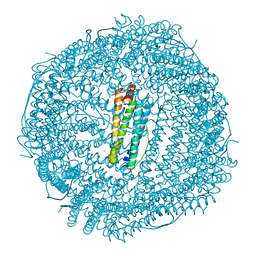 | | Crystal Structure of the Ni-bound Human Heavy-Chain Ferritin 122H-delta C-star variant | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Bailey, J.B, Zhang, L, Chiong, J.A, Ahn, S, Tezcan, F.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Synthetic Modularity of Protein-Metal-Organic Frameworks.
J. Am. Chem. Soc., 139, 2017
|
|
5X3E
 
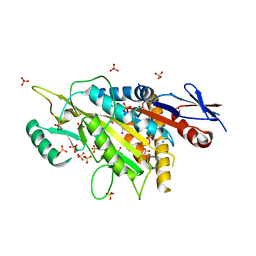 | | kinesin 6 | | Descriptor: | IODIDE ION, Kinesin-like protein, SULFATE ION | | Authors: | Chen, Z, Guan, R, Zhang, L. | | Deposit date: | 2017-02-04 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of Zen4 in the apo state reveals a missing conformation of kinesin
Nat Commun, 8, 2017
|
|
6F0X
 
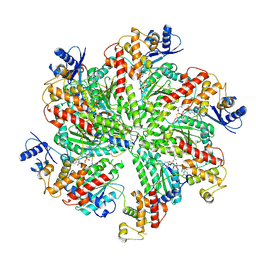 | | Cryo-EM structure of TRIP13 in complex with ATP gamma S, p31comet, C-Mad2 and Cdc20 | | Descriptor: | Cell division cycle protein 20 homolog, MAD2L1-binding protein, Mitotic spindle assembly checkpoint protein MAD2A, ... | | Authors: | Alfieri, C, Chang, L, Barford, D. | | Deposit date: | 2017-11-20 | | Release date: | 2018-05-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism for remodelling of the cell cycle checkpoint protein MAD2 by the ATPase TRIP13.
Nature, 559, 2018
|
|
2RJG
 
 | | Crystal structure of biosynthetic alaine racemase from Escherichia coli | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-10-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
5DP4
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 3 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2S)-2-methyl-3-phenylpropanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
2RJH
 
 | | Crystal structure of biosynthetic alaine racemase in D-cycloserine-bound form from Escherichia coli | | Descriptor: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-10-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
6CSV
 
 | | The structure of the Cep63-Cep152 heterotetrameric complex | | Descriptor: | Centrosomal protein of 63 kDa,Centrosomal protein of 152 kDa | | Authors: | Lee, E, Chen, Y, Zhang, L, Kim, T.S, Ahn, J.I, Park, J.E, Lee, K.S. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of a cylindrical self-assembly at human centrosomes.
Nat Commun, 10, 2019
|
|
5DP9
 
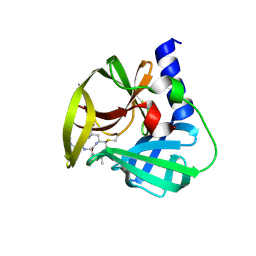 | | Crystal Structure of EV71 3C Proteinase in complex with compound 9 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(cyclobutylmethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP8
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 8 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(2-cyclopropylethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DPA
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 6 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-acetyl-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
