5FHG
 
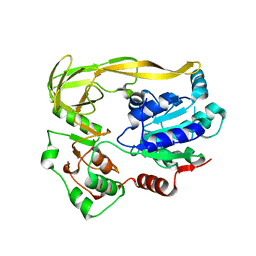 | |
5FHF
 
 | | Crystal structure of Bacteroides sp Pif1 in complex with ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TETRAFLUOROALUMINATE ION, Uncharacterized protein | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
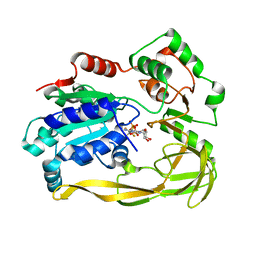
8IND
 
 | | Crystal structure of UGT74AN3-UDP-RES | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, ... | | Authors: | Huang, W. | | Deposit date: | 2023-03-09 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
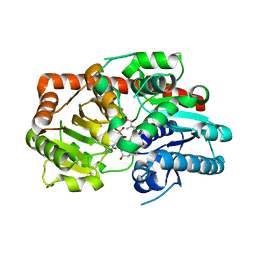
8INV
 
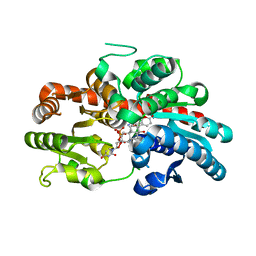 | | Crystal structure of UGT74AN3-UDP-BUF | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
5FL1
 
 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-5-(hydroxymethyl)-2-(prop-2-enylamino)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
8INA
 
 | | Crystal structure of UGT74AN3-UDP | | Descriptor: | GLYCEROL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-09 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8INO
 
 | | Crystal structure of UGT74AN3 in complex UDP and PER | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(3S,5S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-3,5,14-tris(oxidanyl)-2,3,4,6,7,8,9,11,12,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2H-furan-5-one, Glycosyltransferase, ... | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
5FHD
 
 | | Structure of Bacteroides sp Pif1 complexed with tailed dsDNA resulting in ssDNA bound complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*CP*CP*GP*GP*GP*GP*CP*CP*GP*CP*GP*C)-3'), MAGNESIUM ION, ... | | Authors: | Zhou, X, Ren, W, Bharath, S.R, Song, H. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Insights into the Unwinding Mechanism of Bacteroides sp Pif1
Cell Rep, 14, 2016
|
|
5H48
 
 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H4C
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein Cbln4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5GPO
 
 | | The sensor domain structure of the zinc-responsive histidine kinase CzcS from Pseudomonas Aeruginosa | | Descriptor: | SULFATE ION, Sensor protein CzcS, ZINC ION | | Authors: | Wang, D, Chen, W.Z, Huang, S.Q, Liu, X.C, Hu, Q.Y, Wei, T.B, Gan, J.H, Chen, H. | | Deposit date: | 2016-08-03 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structural basis of Zn(II) induced metal detoxification and antibiotic resistance by histidine kinase CzcS in Pseudomonas aeruginosa
PLoS Pathog., 13, 2017
|
|
5H49
 
 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5FL0
 
 | | Structure of a hydrolase with an inhibitor | | Descriptor: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-2-(butylamino)-5-(hydroxymethyl)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d] [1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | Deposit date: | 2015-10-20 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
5GXU
 
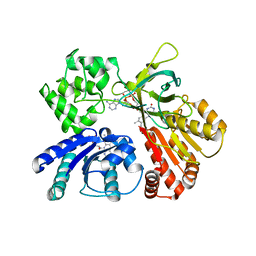 | | Cystal structure of Arabidopsis ATR2 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH--cytochrome P450 reductase 2 | | Authors: | Niu, G, Liu, L. | | Deposit date: | 2016-09-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Arabidopsis thaliana NADPH-cytochrome P450 reductase 2 (ATR2) provides insight into its function
FEBS J., 284, 2017
|
|
3VGX
 
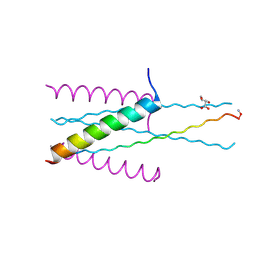 | | Structure of gp41 T21/Cp621-652 | | Descriptor: | ACETIC ACID, Envelope glycoprotein gp160, GLYCEROL | | Authors: | Yao, X, Waltersperger, S, Wang, M, Cui, S. | | Deposit date: | 2011-08-22 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of critical residues for viral entry and inhibition through structural Insight of HIV-1 fusion inhibitor CP621-652.
J.Biol.Chem., 287, 2012
|
|
3VH7
 
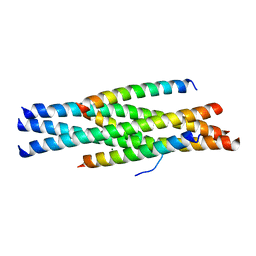 | | Structure of HIV-1 gp41 NHR/fusion inhibitor complex P21 | | Descriptor: | CP32M, Envelope glycoprotein gp160, MAGNESIUM ION | | Authors: | Yao, X, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2011-08-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Structural basis of potent and broad HIV-1 fusion inhibitor CP32M
J.Biol.Chem., 287, 2012
|
|
3VTP
 
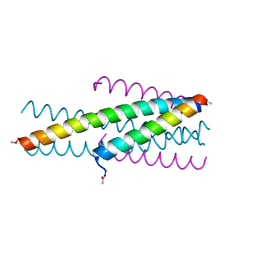 | | HIV fusion inhibitor MT-C34 | | Descriptor: | Transmembrane protein gp41 | | Authors: | Yao, X, Waltersperger, S, Wang, M, Cui, S. | | Deposit date: | 2012-06-02 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The M-T hook structure is critical for design of HIV-1 fusion inhibitors.
J.Biol.Chem., 287, 2012
|
|
3VGY
 
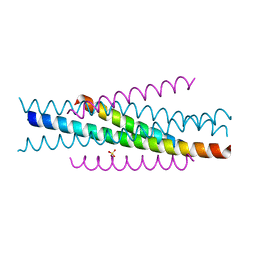 | | Structure of HIV-1 gp41 NHR/fusion inhibitor complex P321 | | Descriptor: | CP32M, Envelope glycoprotein gp160, SULFATE ION | | Authors: | Yao, X, Waltersperger, S, Wang, M.T, Cui, S. | | Deposit date: | 2011-08-22 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | Structural basis of potent and broad HIV-1 fusion inhibitor CP32M
J.Biol.Chem., 287, 2012
|
|
2IJM
 
 | |
6JCS
 
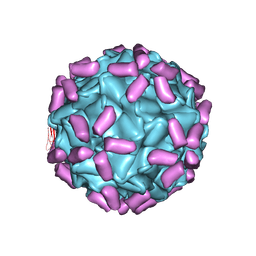 | | AAV5 in complex with AAVR | | Descriptor: | Capsid protein, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-08-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6JCR
 
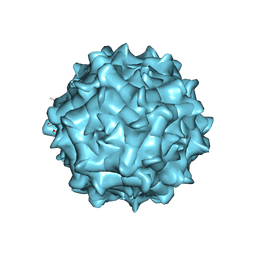 | | AAV1 in neutral condition at 3.07 Ang | | Descriptor: | Capsid protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6JCQ
 
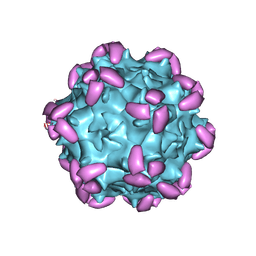 | | AAV1 in complex with AAVR | | Descriptor: | Capsid protein, Dyslexia-associated protein KIAA0319-like protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6JCT
 
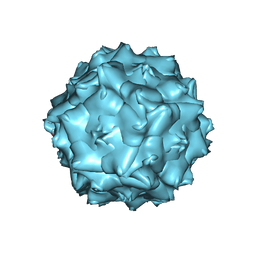 | | AAV5 in neutral condition at 3.18 Ang | | Descriptor: | Capsid protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-07-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
9H8C
 
 | | Human CDK8/Cyclin-C complex with inhibitor 2-9 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3~{S})-3-[2-[(3~{R})-3-fluoranylpyrrolidin-1-yl]pyrimidin-4-yl]piperidin-1-yl]carbonyl-1~{H}-pyrrole-2-carbonitrile, Cyclin-C, ... | | Authors: | Somers, D.O. | | Deposit date: | 2024-10-29 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.573 Å) | | Cite: | Phenotype-Led Identification of IL-10 Upregulators in Human CD4 + T-cells and Elucidation of Their Pharmacology as Highly Selective CDK8/CDK19 Inhibitors.
J.Med.Chem., 68, 2025
|
|
8GCG
 
 | | MDM2 bound to inhibitor | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, macrocyclic peptide inhibitor | | Authors: | Silvestri, A.P, Muir, E.W, Chakka, S.K, Tripathi, S.M, Rubin, S.M, Pye, C.R, Schwochert, J.A. | | Deposit date: | 2023-03-01 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | DNA-Encoded Macrocyclic Peptide Libraries Enable the Discovery of a Neutral MDM2-p53 Inhibitor.
Acs Med.Chem.Lett., 14, 2023
|
|
