6WMI
 
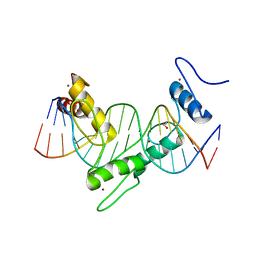 | | ZNF410 zinc fingers 1-5 with 17 mer blunt DNA Oligonucleotide | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*CP*AP*TP*CP*CP*CP*AP*TP*AP*AP*TP*AP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*TP*AP*TP*GP*GP*GP*AP*TP*GP*TP*G)-3'), ... | | Authors: | Ren, R, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | ZNF410 Uniquely Activates the NuRD Component CHD4 to Silence Fetal Hemoglobin Expression.
Mol.Cell, 81, 2021
|
|
7TXC
 
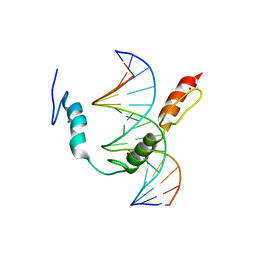 | | HIC2 zinc finger domain in complex with the DNA binding motif-2 of the BCL11A enhancer | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*TP*GP*GP*CP*AP*TP*TP*AP*TP*CP*T)-3'), DNA (5'-D(*AP*GP*AP*TP*AP*AP*TP*GP*CP*CP*AP*AP*CP*AP*GP*T)-3'), Hypermethylated in cancer 2 protein, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | HIC2 controls developmental hemoglobin switching by repressing BCL11A transcription.
Nat.Genet., 54, 2022
|
|
8THF
 
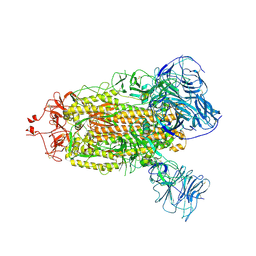 | | SARS-CoV-2 BA.1 S-6P-no-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | Authors: | Bu, F, Li, F, Liu, B. | | Deposit date: | 2023-07-15 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Universal subunit vaccine protects against multiple SARS-CoV-2 variants and SARS-CoV.
Npj Vaccines, 9, 2024
|
|
5CA9
 
 | |
5CA8
 
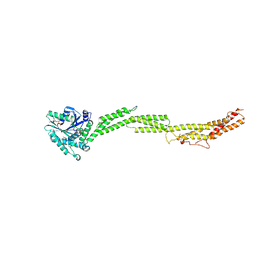 | |
6LJJ
 
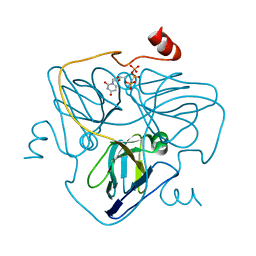 | | Swine dUTPase in complex with alpha,beta-iminodUTP and magnesium ion | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, mitochondrial isoform 1, ... | | Authors: | Liang, R, Peng, G.Q. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural comparisons of host and African swine fever virus dUTPases reveal new clues for inhibitor development.
J.Biol.Chem., 296, 2020
|
|
8YW6
 
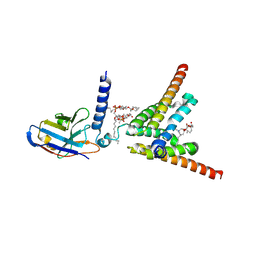 | | Cryo-EM structure of apo human mitochondrial pyruvate carrier in the IMS-open conformation at pH 8.0 | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, MPC specific nanobody 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-03-29 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
8YW9
 
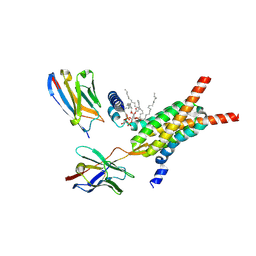 | | Cryo-EM structure of human mitochondrial pyruvate carrier in the matrix-facing conformation at pH 6.8 | | Descriptor: | CARDIOLIPIN, MPC specific nanobody 1, MPC specific nanobody 2, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-03-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
8YW8
 
 | | Cryo-EM structure of human mitochondrial pyruvate carrier in complex with the inhibitor UK5099 | | Descriptor: | (E)-2-cyano-3-(1-phenylindol-3-yl)prop-2-enoic acid, CARDIOLIPIN, MPC specific nanobody 1, ... | | Authors: | Shi, J.H, Liang, J.M, Ma, D. | | Deposit date: | 2024-03-30 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structures and mechanism of the human mitochondrial pyruvate carrier.
Nature, 641, 2025
|
|
7CYC
 
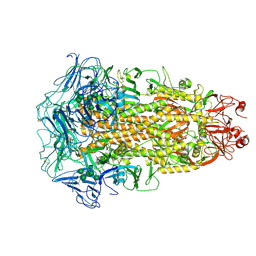 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
7CYD
 
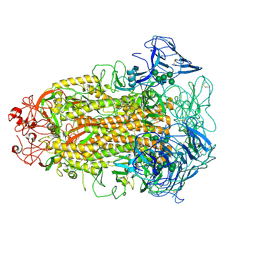 | | Cryo-EM structures of Alphacoronavirus spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, X, Shi, Y, Ding, W, Liu, Z.J, Peng, G. | | Deposit date: | 2020-09-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Cryo-EM analysis of the HCoV-229E spike glycoprotein reveals dynamic prefusion conformational changes.
Nat Commun, 12, 2021
|
|
4P0N
 
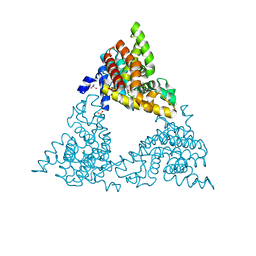 | | Crystal structure of PDE10a with a novel Imidazo[4,5-b]pyridine inhibitor | | Descriptor: | GLYCEROL, N-[cis-3-(2-methoxy-3H-imidazo[4,5-b]pyridin-3-yl)cyclobutyl]-1,3-benzothiazol-2-amine, N-[trans-3-(2-methoxy-3H-imidazo[4,5-b]pyridin-3-yl)cyclobutyl]-1,3-benzothiazol-2-amine, ... | | Authors: | Chmait, S. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of Novel Imidazo[4,5-b]pyridines as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
Acs Med.Chem.Lett., 5, 2014
|
|
4MUW
 
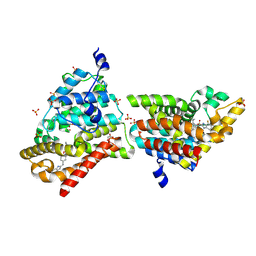 | | Crystal Structure of PDE10A with Novel Keto-Benzimidazole Inhibitor | | Descriptor: | 2-{4-[(6,7-difluoro-1H-benzimidazol-2-yl)amino]phenoxy}-N-methyl-3,4'-bipyridin-2'-amine, GLYCEROL, SULFATE ION, ... | | Authors: | Chmait, S, Jordan, S. | | Deposit date: | 2013-09-23 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.639 Å) | | Cite: | Design, Optimization, and Biological Evaluation of Novel Keto-Benzimidazoles as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
J.Med.Chem., 56, 2013
|
|
4MVH
 
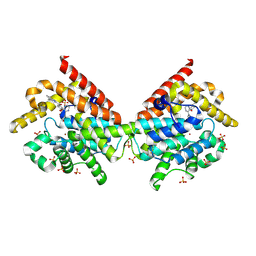 | | Crystal Structure of PDE10A with Novel Keto-Benzimidazole Inhibitor | | Descriptor: | 1H-benzimidazol-2-yl(4-{[3-(morpholin-4-yl)pyrazin-2-yl]oxy}phenyl)methanone, SULFATE ION, ZINC ION, ... | | Authors: | Chmait, S, Jordan, S. | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Design, Optimization, and Biological Evaluation of Novel Keto-Benzimidazoles as Potent and Selective Inhibitors of Phosphodiesterase 10A (PDE10A).
J.Med.Chem., 56, 2013
|
|
8H8J
 
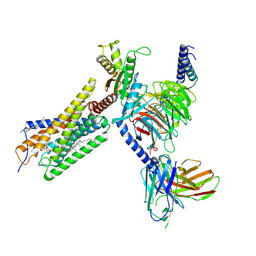 | | Lodoxamide-bound GPR35 in complex with G13 | | Descriptor: | 2,2'-[(2-chloro-5-cyano-1,3-phenylene)bis(azanediyl)]bis(oxoacetic acid), CALCIUM ION, CHOLESTEROL, ... | | Authors: | Yuan, Q, Duan, J, Liu, Q, Xu, H.E, Jiang, Y. | | Deposit date: | 2022-10-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Insights into divalent cation regulation and G 13 -coupling of orphan receptor GPR35.
Cell Discov, 8, 2022
|
|
4DG6
 
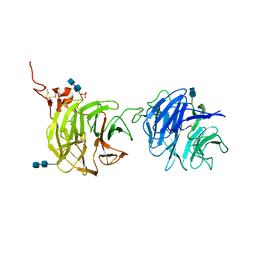 | | Crystal structure of domains 1 and 2 of LRP6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Low-density lipoprotein receptor-related protein 6, PHOSPHATE ION | | Authors: | Ceska, T.A, Doyle, C, Slocombe, P. | | Deposit date: | 2012-01-25 | | Release date: | 2012-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Characterization of the interaction of sclerostin with the LRP family of Wnt co-receptors
To be Published
|
|
8I6O
 
 | |
8I6R
 
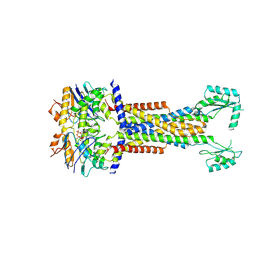 | | Cryo-EM structure of Pseudomonas aeruginosa FtsE(E163Q)X/EnvC complex with ATP in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Xu, X, Li, J, Luo, M. | | Deposit date: | 2023-01-29 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanistic insights into the regulation of cell wall hydrolysis by FtsEX and EnvC at the bacterial division site.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8I6S
 
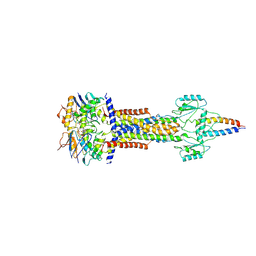 | | Cryo-EM structure of Pseudomonas aeruginosa FtsE(E163Q)X/EnvC complex with ATP in peptidisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Xu, X, Li, J, Luo, M. | | Deposit date: | 2023-01-29 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanistic insights into the regulation of cell wall hydrolysis by FtsEX and EnvC at the bacterial division site.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
9KGP
 
 | |
8JB0
 
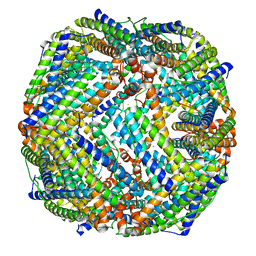 | |
8JAX
 
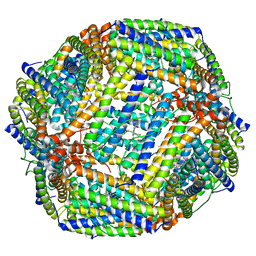 | |
8W6F
 
 | | Apo structure of HBC binder | | Descriptor: | wFAP1.1 structure | | Authors: | Liang, M.F, Zhu, J.Y, Lu, P.L. | | Deposit date: | 2023-08-28 | | Release date: | 2024-11-27 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | De novo design of transmembrane fluorescence-activating proteins.
Nature, 640, 2025
|
|
8W6E
 
 | | De novo design of HBC599 binder | | Descriptor: | 4-[(Z)-1-cyano-2-{6-[(2-hydroxyethyl)(methyl)amino]-1-benzothiophen-2-yl}ethenyl]benzonitrile, HBC599 in complex with wFAP1.1 | | Authors: | Liang, M.F, Zhu, J.Y, Lu, P.L. | | Deposit date: | 2023-08-28 | | Release date: | 2024-11-27 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | De novo design of transmembrane fluorescence-activating proteins.
Nature, 640, 2025
|
|
8WIL
 
 | | Crystal structure of Jingmen tick virus RNA-dependent RNA polymerase (D55 construct) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Jingmen tick virus NSP1, ... | | Authors: | Wang, X, Jing, X, Deng, F, Gong, P. | | Deposit date: | 2023-09-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A jingmenvirus RNA-dependent RNA polymerase structurally resembles the flavivirus counterpart but with different features at the initiation phase.
Nucleic Acids Res., 52, 2024
|
|
