6OO5
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in amphipol resolved to 4.2 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
6OO3
 
 | | Cryo-EM structure of the C4-symmetric TRPV2/RTx complex in amphipol resolved to 2.9 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
6OO4
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in amphipol resolved to 3.3 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
6NR3
 
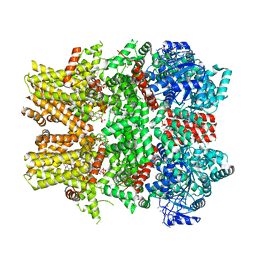 | | Cryo-EM structure of the TRPM8 ion channel in complex with high occupancy icilin, PI(4,5)P2, and calcium | | Descriptor: | (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, CALCIUM ION, Icilin, ... | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
6OO7
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in nanodiscs | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
6OB6
 
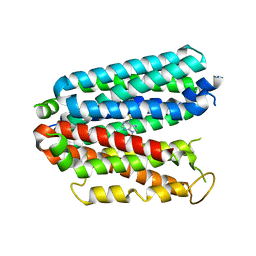 | |
6OB7
 
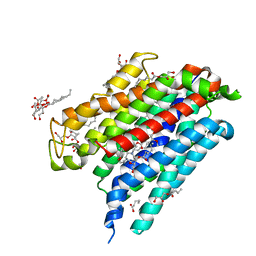 | | Human equilibrative nucleoside transporter-1, dilazep bound | | Descriptor: | (1,4-diazepane-1,4-diyl)di(propane-3,1-diyl) bis(3,4,5-trimethoxybenzoate), (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Equilibrative nucleoside transporter 1 | | Authors: | Wright, N.J, Lee, S.-Y. | | Deposit date: | 2019-03-19 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of human ENT1 in complex with adenosine reuptake inhibitors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2X26
 
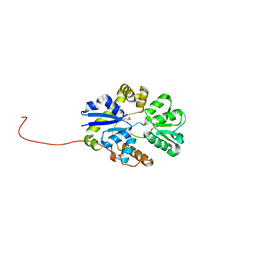 | | Crystal structure of the periplasmic aliphatic sulphonate binding protein SsuA from Escherichia coli | | Descriptor: | GLYCEROL, PERIPLASMIC ALIPHATIC SULPHONATES-BINDING PROTEIN | | Authors: | Beale, J, Lee, S, Iwata, S, Beis, K. | | Deposit date: | 2010-01-11 | | Release date: | 2010-04-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Aliphatic Sulfonate-Binding Protein Ssua from Escherichia Coli
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6OT2
 
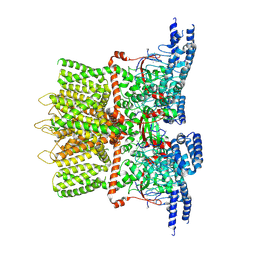 | | Structure of the TRPV3 K169A sensitized mutant in apo form at 4.1 A resolution | | Descriptor: | Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Borschel, W.F, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Regulatory switch at the cytoplasmic interface controls TRPV channel gating.
Elife, 8, 2019
|
|
6PKW
 
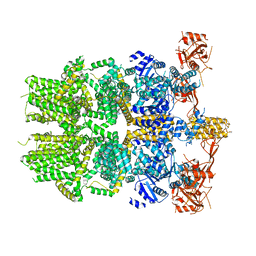 | | Cryo-EM structure of the zebrafish TRPM2 channel in the apo conformation, processed with C2 symmetry (pseudo C4 symmetry) | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yin, Y, Wu, M, Hsu, A.L, Borschel, W.F, Borgnia, M.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
6OT5
 
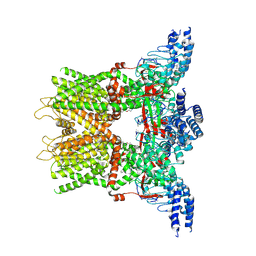 | | Structure of the TRPV3 K169A sensitized mutant in the presence of 2-APB at 3.6 A resolution | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 3,Transient receptor potential cation channel subfamily V member 3 | | Authors: | Zubcevic, L, Borschel, W.F, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-05-02 | | Release date: | 2019-05-22 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Regulatory switch at the cytoplasmic interface controls TRPV channel gating.
Elife, 8, 2019
|
|
6PKV
 
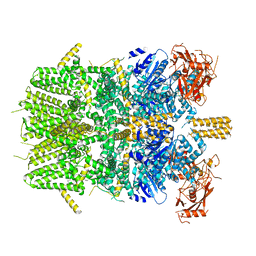 | | Cryo-EM structure of the zebrafish TRPM2 channel in the apo conformation, processed with C4 symmetry | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yin, Y, Wu, M, Hsu, A.L, Borschel, W.F, Borgnia, M.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
3QMX
 
 | |
5XJH
 
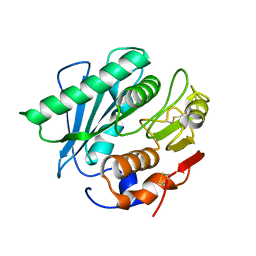 | |
7STL
 
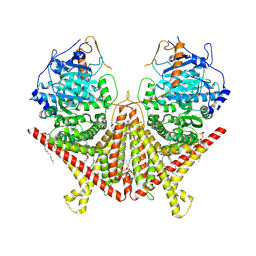 | | Chitin Synthase 2 from Candida albicans at the apo state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Chitin synthase | | Authors: | Ren, Z, Chhetri, A, Lee, S, Yokoyama, K. | | Deposit date: | 2021-11-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for inhibition and regulation of a chitin synthase from Candida albicans.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TX7
 
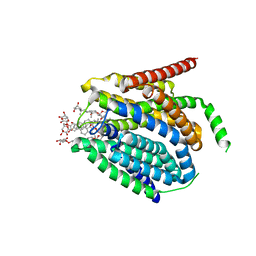 | |
7TX6
 
 | | Cryo-EM structure of the human reduced folate carrier in complex with methotrexate | | Descriptor: | Digitonin, METHOTREXATE, Reduced folate transporter,Soluble cytochrome b562 | | Authors: | Wright, N.J, Fedor, J.G, Lee, S.-Y. | | Deposit date: | 2022-02-07 | | Release date: | 2022-09-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Methotrexate recognition by the human reduced folate carrier SLC19A1.
Nature, 609, 2022
|
|
3TIJ
 
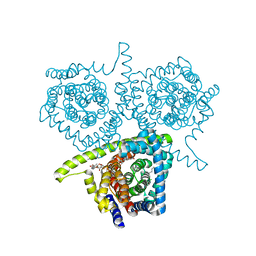 | | Crystal structure of a concentrative nucleoside transporter from Vibrio cholerae | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, SODIUM ION, ... | | Authors: | Johnson, Z.L, Cheong, C.-G, Lee, S.-Y. | | Deposit date: | 2011-08-20 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Crystal structure of a concentrative nucleoside transporter from Vibrio cholerae at 2.4A
Nature, 483, 2012
|
|
1BLI
 
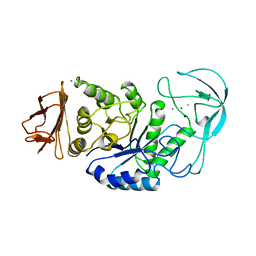 | | BACILLUS LICHENIFORMIS ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Machius, M, Declerck, N, Huber, R, Wiegand, G. | | Deposit date: | 1998-01-07 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Activation of Bacillus licheniformis alpha-amylase through a disorder-->order transition of the substrate-binding site mediated by a calcium-sodium-calcium metal triad.
Structure, 6, 1998
|
|
2QR8
 
 | |
2QR7
 
 | |
5K48
 
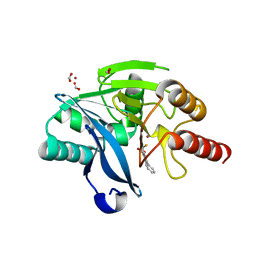 | | VIM-2 Metallo Beta Lactamase in complex with 3-(mercaptomethyl)-[1,1'-biphenyl]-4-carboxylic acid | | Descriptor: | 4-phenyl-2-(sulfanylmethyl)benzoic acid, Beta-lactamase VIM-2, FORMIC ACID, ... | | Authors: | Zollman, D, McDonough, M, Brem, J, Schofield, C. | | Deposit date: | 2016-05-20 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | In Silico Fragment-Based Design Identifies Subfamily B1 Metallo-beta-lactamase Inhibitors.
J. Med. Chem., 61, 2018
|
|
5AHR
 
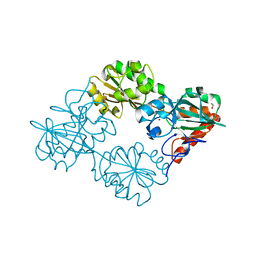 | | Crystal structure of human DNA cross-link repair 1A, crystal form B | | Descriptor: | 1,2-ETHANEDIOL, DNA CROSS-LINK REPAIR 1A PROTEIN, ZINC ION | | Authors: | Allerston, C.K, Newman, J.A, Vollmar, M, Goubin, S, Forese, D.S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2015-02-06 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Structures of the Snm1A and Snm1B/Apollo Nuclease Domains Reveal a Potential Basis for Their Distinct DNA Processing Activities.
Nucleic Acids Res., 43, 2015
|
|
5B6C
 
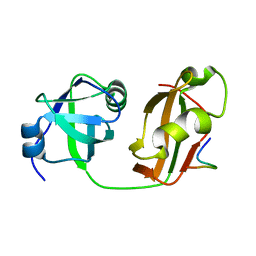 | | Structural Details of Ufd1 binding to p97 | | Descriptor: | Peptide from Ubiquitin fusion degradation protein 1 homolog, Transitional endoplasmic reticulum ATPase | | Authors: | Le, L.T.M, Yang, J.K. | | Deposit date: | 2016-05-26 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Details of Ufd1 Binding to p97 and Their Functional Implications in ER-Associated Degradation
PLoS ONE, 11, 2016
|
|
5AHO
 
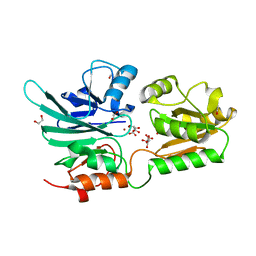 | | Crystal structure of human 5' exonuclease Apollo | | Descriptor: | 1,2-ETHANEDIOL, 5' EXONUCLEASE APOLLO, L(+)-TARTARIC ACID, ... | | Authors: | Allerston, C.K, Vollmar, M, Krojer, T, Pike, A.C.W, Newman, J.A, Carpenter, E, Quigley, A, Mahajan, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Gileadi, O. | | Deposit date: | 2015-02-06 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The Structures of the Snm1A and Snm1B/Apollo Nuclease Domains Reveal a Potential Basis for Their Distinct DNA Processing Activities.
Nucleic Acids Res., 43, 2015
|
|
