4H75
 
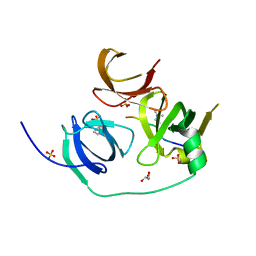 | | Crystal structure of human Spindlin1 in complex with a histone H3K4(me3) peptide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Histone H3, ... | | Authors: | Yang, N, Wang, W, Wang, Y, Wang, M, Zhao, Q, Rao, Z, Zhu, B, Xu, R.M. | | Deposit date: | 2012-09-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1VIF
 
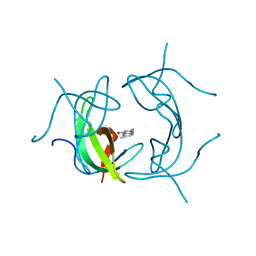 | | STRUCTURE OF DIHYDROFOLATE REDUCTASE | | Descriptor: | DIHYDROFOLATE REDUCTASE, FOLIC ACID | | Authors: | Narayana, N, Matthews, D.A, Howell, E.E, Xuong, N.-H. | | Deposit date: | 1996-10-03 | | Release date: | 1997-10-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A plasmid-encoded dihydrofolate reductase from trimethoprim-resistant bacteria has a novel D2-symmetric active site.
Nat.Struct.Biol., 2, 1995
|
|
1D28
 
 | |
5B1A
 
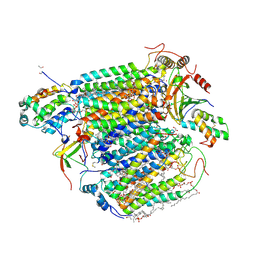 | | Bovine heart cytochrome c oxidase in the fully oxidized state at 1.5 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Yano, N, Muramoto, K, Shimada, A, Takemura, S, Baba, J, Fujisawa, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2015-12-01 | | Release date: | 2016-09-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Mg2+-containing Water Cluster of Mammalian Cytochrome c Oxidase Collects Four Pumping Proton Equivalents in Each Catalytic Cycle.
J.Biol.Chem., 291, 2016
|
|
8TCI
 
 | | Crystal structure of DNMT3C-DNMT3L in complex with CGG DNA | | Descriptor: | (5'-D(P*CP*AP*TP*G)-R(P*(PYO))-D(P*GP*GP*TP*CP*TP*AP*AP*TP*TP*AP*GP*AP*CP*CP*GP*CP*AP*TP*G)-3'), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3C, ... | | Authors: | Khudaverdyan, N, Song, J. | | Deposit date: | 2023-07-01 | | Release date: | 2024-08-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The structure of DNA methyltransferase DNMT3C reveals an activity-tuning mechanism for DNA methylation.
J.Biol.Chem., 300, 2024
|
|
2F8W
 
 | | Crystal structure of d(CACGTG)2 | | Descriptor: | 1,3-DIAMINOPROPANE, 5'-D(*CP*AP*CP*GP*TP*G)-3', SPERMINE | | Authors: | Narayana, N, Shamala, N, Ganesh, K.N, Viswamitra, M.A. | | Deposit date: | 2005-12-04 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Interaction between the Z-Type DNA Duplex and 1,3-Propanediamine: Crystal Structure of d(CACGTG)2 at 1.2 A Resolution
Biochemistry, 45, 2006
|
|
7YQS
 
 | | Neutron structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-23 | | Method: | NEUTRON DIFFRACTION (1.25 Å), X-RAY DIFFRACTION | | Cite: | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
3X2Q
 
 | | X-ray structure of cyanide-bound bovine heart cytochrome c oxidase in the fully oxidized state at 2.0 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Yano, N, Muramoto, K, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2014-12-26 | | Release date: | 2015-06-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of cyanide-bound bovine heart cytochrome c oxidase in the fully oxidized state at 2.0 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
2GQV
 
 | |
8I4D
 
 | | X-ray structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
2ZGK
 
 | | Crystal structure of wildtype AAL | | Descriptor: | Anti-tumor lectin | | Authors: | Yang, N, Li, D.F, Wang, D.C. | | Deposit date: | 2008-01-23 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the tumor cell apoptosis-inducing activity of an antitumor lectin from the edible mushroom Agrocybe aegerita
J.Mol.Biol., 387, 2009
|
|
8FZ9
 
 | |
2ZGR
 
 | |
2ZGN
 
 | | crystal structure of recombinant Agrocybe aegerita lectin, rAAL, complex with galactose | | Descriptor: | Anti-tumor lectin, beta-D-galactopyranose | | Authors: | Yang, N, Li, D.F, Wang, D.C. | | Deposit date: | 2008-01-23 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the tumor cell apoptosis-inducing activity of an antitumor lectin from the edible mushroom Agrocybe aegerita
J.Mol.Biol., 387, 2009
|
|
2ZGP
 
 | |
2ZGL
 
 | |
2B1L
 
 | |
2GYP
 
 | | Diabetes mellitus due to a frustrated Schellman motif in HNF-1a | | Descriptor: | Hepatocyte nuclear factor 1-alpha | | Authors: | Narayana, N, Phillips, N.B, Hua, Q.X, Jia, W, Weiss, M.A. | | Deposit date: | 2006-05-09 | | Release date: | 2007-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diabetes mellitus due to misfolding of a beta-cell transcription factor: stereospecific frustration of a Schellman motif in HNF-1alpha.
J.Mol.Biol., 362, 2006
|
|
2KZ9
 
 | |
6K0W
 
 | | DNA methyltransferase in complex with sinefungin | | Descriptor: | Adenine specific DNA methyltransferase (Mod), SINEFUNGIN | | Authors: | Narayanan, N, Nair, D.T. | | Deposit date: | 2019-05-07 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Tetramerization at Low pH Licenses DNA Methylation Activity of M.HpyAXI in the Presence of Acid Stress.
J.Mol.Biol., 432, 2020
|
|
2B1K
 
 | | Crystal structure of E. coli CcmG protein | | Descriptor: | Thiol:disulfide interchange protein dsbE | | Authors: | Ouyang, N, Gao, Y.G, Hu, H.Y, Xia, Z.X. | | Deposit date: | 2005-09-15 | | Release date: | 2006-09-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of E. coli CcmG and its mutants reveal key roles of the N-terminal beta-sheet and the fingerprint region
Proteins, 65, 2006
|
|
3A4O
 
 | | Lyn kinase domain | | Descriptor: | STAUROSPORINE, Tyrosine-protein kinase Lyn | | Authors: | Miyano, N, Kinoshita, T, Tada, T. | | Deposit date: | 2009-07-11 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitor recognition of human Lyn kinase domain
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6P2J
 
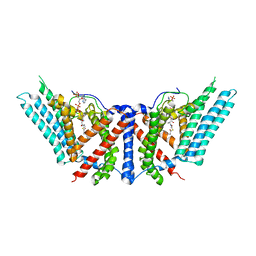 | | Dimeric structure of ACAT1 | | Descriptor: | S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name), Sterol O-acyltransferase 1 | | Authors: | Yan, N, Qian, H.W. | | Deposit date: | 2019-05-21 | | Release date: | 2020-05-20 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for catalysis and substrate specificity of human ACAT1.
Nature, 581, 2020
|
|
6P2P
 
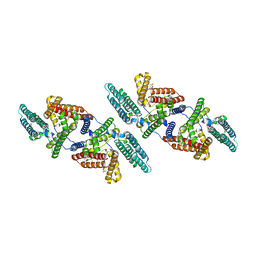 | | Tetrameric structure of ACAT1 | | Descriptor: | S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name), Sterol O-acyltransferase 1 | | Authors: | Yan, N, Qian, H.W. | | Deposit date: | 2019-05-21 | | Release date: | 2020-05-20 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for catalysis and substrate specificity of human ACAT1.
Nature, 581, 2020
|
|
6W5S
 
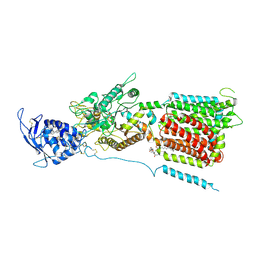 | | NPC1 structure in GDN micelles at pH 8.0 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
