6ZHT
 
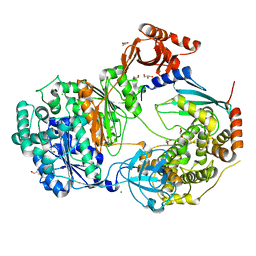 | | Uba1-Ubc13 disulfide mediated complex | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Schaefer, A, Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system.
To Be Published
|
|
6ZHU
 
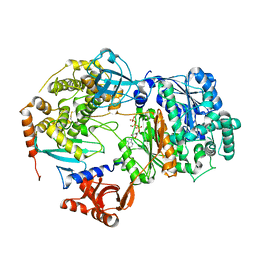 | | Yeast Uba1 in complex with Ubc3 and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2020-06-23 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | ATP induced conformational changes facilitate E1-E2 disulfide bridging in the ubiquitin system
To Be Published
|
|
1QZT
 
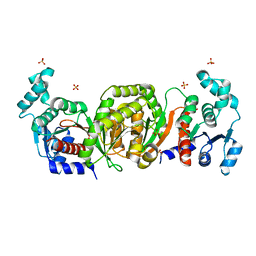 | | Phosphotransacetylase from Methanosarcina thermophila | | Descriptor: | Phosphate acetyltransferase, SULFATE ION | | Authors: | Iyer, P.P, Lawrence, S.H, Luther, K.B, Rajashankar, K.R, Yennawar, H.P, Ferry, J.G, Schindelin, H. | | Deposit date: | 2003-09-17 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of phosphotransacetylase from the methanogenic archaeon Methanosarcina thermophila.
STRUCTURE, 12, 2004
|
|
5AES
 
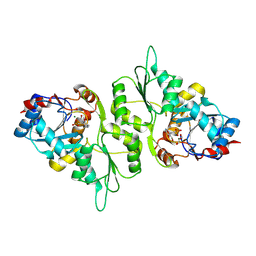 | | Crystal Structure of murine Chronophin (Pyridoxal Phosphate Phosphatase) in Complex with a PNP-derived Inhibitor | | Descriptor: | GLYCEROL, MAGNESIUM ION, PYRIDOXAL PHOSPHATE PHOSPHATASE, ... | | Authors: | Knobloch, G, Jabari, N, Koehn, M, Gohla, A, Schindelin, H. | | Deposit date: | 2015-01-09 | | Release date: | 2015-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Synthesis of Hydrolysis-Resistant Pyridoxal 5'-Phosphate Analogs and Their Biochemical and X-Ray Crystallographic Characterization with the Pyridoxal Phosphatase Chronophin.
Bioorg.Med.Chem., 23, 2015
|
|
7ZH9
 
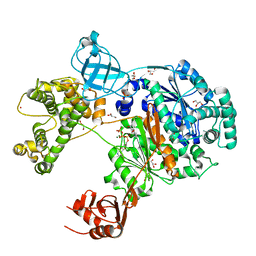 | | Uba1 in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
7ZTL
 
 | |
8A1I
 
 | |
3TIW
 
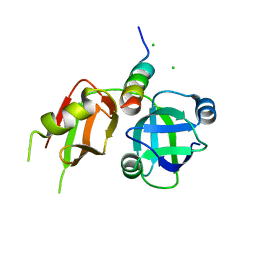 | | Crystal structure of p97N in complex with the C-terminus of gp78 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase AMFR, Transitional endoplasmic reticulum ATPase | | Authors: | Haenzelmann, P, Schindelin, H. | | Deposit date: | 2011-08-22 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | The Structural and Functional Basis of the p97/Valosin-containing Protein (VCP)-interacting Motif (VIM): MUTUALLY EXCLUSIVE BINDING OF COFACTORS TO THE N-TERMINAL DOMAIN OF p97.
J.Biol.Chem., 286, 2011
|
|
5L6H
 
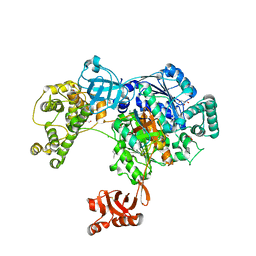 | | Uba1 in complex with Ub-ABPA3 covalent adduct | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
5L6I
 
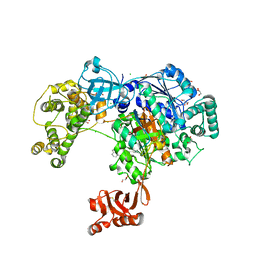 | | Uba1 in complex with Ub-MLN4924 covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
5L6J
 
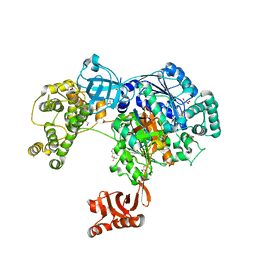 | | Uba1 in complex with Ub-MLN7243 covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
3ESW
 
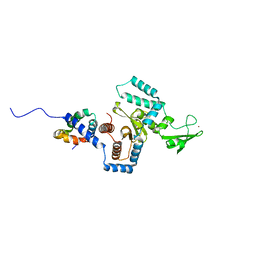 | | Complex of yeast PNGase with GlcNAc2-IAc. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase, UV excision repair protein RAD23, ... | | Authors: | Zhao, G, Zhou, X, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and mutational studies on the importance of oligosaccharide binding for the activity of yeast PNGase.
Glycobiology, 19, 2009
|
|
3CMM
 
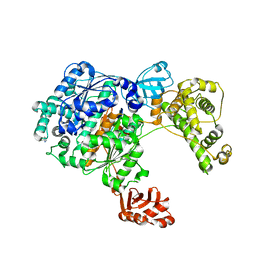 | | Crystal Structure of the Uba1-Ubiquitin Complex | | Descriptor: | PROLINE, Ubiquitin, Ubiquitin-activating enzyme E1 1 | | Authors: | Lee, I, Schindelin, H. | | Deposit date: | 2008-03-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes.
Cell(Cambridge,Mass.), 134, 2008
|
|
3M62
 
 | |
3M63
 
 | |
4F9Z
 
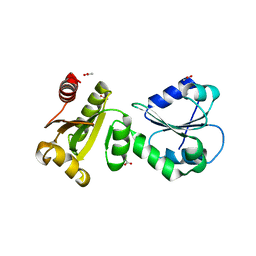 | | Crystal Structure of human ERp27 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Kober, F.X, Koelmel, W, Kuper, J, Schindelin, H. | | Deposit date: | 2012-05-21 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Protein-Disulfide Isomerase Family Member ERp27 Provides Insights into Its Substrate Binding Capabilities.
J.Biol.Chem., 288, 2013
|
|
6YK0
 
 | |
6YJZ
 
 | |
6YK1
 
 | |
3IPO
 
 | | Crystal structure of YnjE | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3IPP
 
 | | crystal structure of sulfur-free YnjE | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative thiosulfate sulfurtransferase ynjE, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3GAE
 
 | | Crystal Structure of PUL | | Descriptor: | CHLORIDE ION, GLYCEROL, Protein DOA1 | | Authors: | Zhao, G, Schindelin, H, Lennarz, W.J. | | Deposit date: | 2009-02-17 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Armadillo motif in Ufd3 interacts with Cdc48 and is involved in ubiquitin homeostasis and protein degradation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3QQ7
 
 | |
3R3M
 
 | |
3QQ8
 
 | |
