8OWT
 
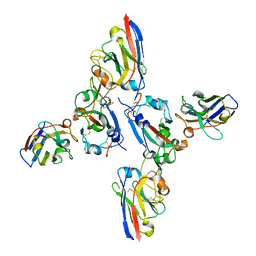 | | SARS-CoV-2 spike RBD with A8 and H3 nanobodies bound | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A8, ... | | Authors: | Mikolajek, H, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWW
 
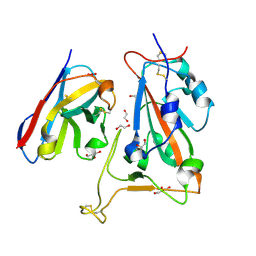 | | B5-5 nanobody bound to SARS-CoV-2 spike RBD (Wuhan) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, B5-5 nanobody, ... | | Authors: | Cornish, K.A.S, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OWV
 
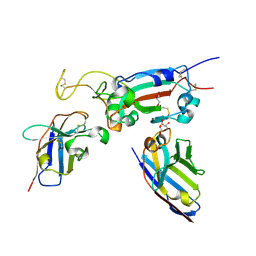 | | H6 and F2 nanobodies bound to SARS-CoV-2 spike RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2, GLYCEROL, ... | | Authors: | Mikolajek, H, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OYU
 
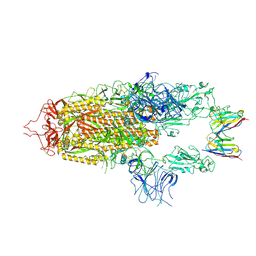 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '2 up 1 down' RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | Authors: | Weckener, M, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
8OYT
 
 | | Stabilised BA.1 SARS-CoV-2 spike with H6 nanobodies in '3 up' RBD conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H6 nanobody, ... | | Authors: | Weckener, M, Naismith, J.H, Owens, R.J. | | Deposit date: | 2023-05-05 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and functional characterization of nanobodies that neutralize Omicron variants of SARS-CoV-2.
Open Biology, 14, 2024
|
|
5DF7
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH AZLOCILLIN | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-2-oxo-1-{[(2R)-2-{[(2-oxoimidazolidin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}ethyl]-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Cell division protein, ... | | Authors: | Ren, J, Nettleship, J.E, Males, A, Stuart, D.I, Owens, R.J. | | Deposit date: | 2015-08-26 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 in complexes with azlocillin and cefoperazone in both acylated and deacylated forms.
Febs Lett., 590, 2016
|
|
6QNA
 
 | | Structure of bovine anti-RSV hybrid Fab B13HC-B4LC | | Descriptor: | B13 Heavy chain, B4 light chain, GLYCEROL | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QN9
 
 | | Structure of bovine anti-RSV Fab B4 | | Descriptor: | GLYCEROL, Heavy chain, SULFATE ION, ... | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
6QN8
 
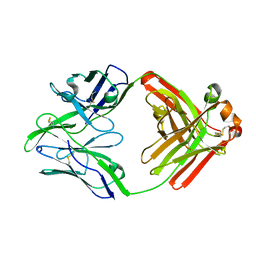 | | Structure of bovine anti-RSV Fab B13 | | Descriptor: | CHLORIDE ION, Heavy chain of bovine anti-RSV B13 Fab, Light chain of bovine anti-RSV Fab B13 | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
5EYV
 
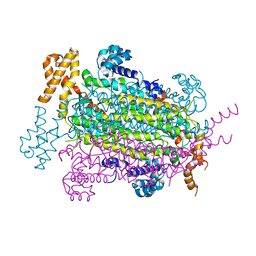 | | Crystal Structure of Adenylosuccinate lyase from Schistosoma mansoni in APO form. | | Descriptor: | Adenylosuccinate lyase | | Authors: | Romanello, L, Torini, J.R, Bird, L.E, Nettleship, J.E, Owens, R.J, Reddivari, Y, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural and kinetic analysis of Schistosoma mansoni Adenylosuccinate Lyase (SmADSL).
Mol. Biochem. Parasitol., 214, 2017
|
|
6QN7
 
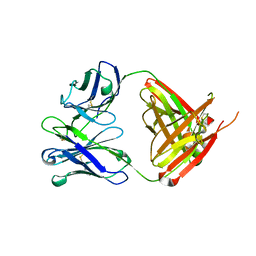 | | Structure of bovine anti-RSV hybrid Fab B4HC-B13LC | | Descriptor: | Heavy chain of bovine anti-RSV B4, Light chain of bovine anti-RSV B13 | | Authors: | Ren, J, Nettleship, J.E, Harris, G, Mwangi, W, Rhaman, N, Grant, C, Kotecha, A, Fry, E, Charleston, B, Stuart, D.I, Hammond, J, Owens, R.J. | | Deposit date: | 2019-02-10 | | Release date: | 2019-05-29 | | Last modified: | 2019-08-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The role of the light chain in the structure and binding activity of two cattle antibodies that neutralize bovine respiratory syncytial virus.
Mol.Immunol., 112, 2019
|
|
8P6H
 
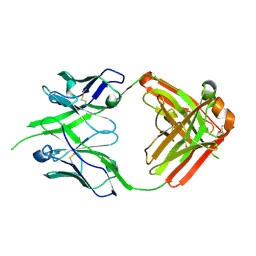 | | Bovine naive ultralong antibody AbBLV5B8* collected at 100K | | Descriptor: | Antibody BLV5B8* heavy chain, Antibody BLV5B8* light chain | | Authors: | Clarke, J.D, Douangamath, A, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2023-05-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The impact of chain-exchange on bovine ultralong immunoglobulins.
Acta Crystallographica Section F, 2023
|
|
8P2T
 
 | | Bovine naive ultralong antibody AbD08* collected at 100K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Antibody D08* heavy chain, Antibody D08* light chain, ... | | Authors: | Clarke, J.D, Douangamath, A, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2023-05-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The impact of chain-exchange on bovine ultralong immunoglobulins.
Acta Crystallographica Section F, 2024
|
|
4BBK
 
 | | Structural and functional characterisation of the kindlin-1 pleckstrin homology domain | | Descriptor: | FERMITIN FAMILY HOMOLOG 1, GLYCEROL | | Authors: | Yates, L.A, Lumb, C.N, Brahme, N.N, Zalyte, R, Bird, L.E, De Colibus, L, Owens, R.J, Calderwood, D.A, Sansom, M.S.P, Gilbert, R.J.C. | | Deposit date: | 2012-09-25 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterisation of the Kindlin-1 Pleckstrin Homology Domain
J.Biol.Chem., 287, 2012
|
|
8PDD
 
 | | Thioredoxin glutathione reductase of Schistosoma mansoni at 1.25A resolution. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ribeiro, L, Montoya, B.O, Moreira-Filho, J.T, Bowyer, S, Verma, A, Neves, B.J, Owens, R.J, Andrade, C.H, Silva-Jr, F.P, Furnham, N. | | Deposit date: | 2023-06-12 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Fragment library screening by X-ray crystallography and binding site analysis on thioredoxin glutathione reductase of Schistosoma mansoni.
Sci Rep, 14, 2024
|
|
3HHF
 
 | | Structure of CrgA regulatory domain, a LysR-type transcriptional regulator from Neisseria meningitidis. | | Descriptor: | CHLORIDE ION, Transcriptional regulator, LysR family | | Authors: | Sainsbury, S, Ren, J, Owens, R.J, Stuart, D.I, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-05-15 | | Release date: | 2009-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators
Nucleic Acids Res., 37, 2009
|
|
3HHG
 
 | | Structure of CrgA, a LysR-type transcriptional regulator from Neisseria meningitidis. | | Descriptor: | Transcriptional regulator, LysR family | | Authors: | Sainsbury, S, Ren, J, Owens, R.J, Stuart, D.I, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-05-15 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators
Nucleic Acids Res., 37, 2009
|
|
2Y7Q
 
 | | THE HIGH-AFFINITY COMPLEX BETWEEN IGE AND ITS RECEPTOR FC EPSILON RI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIGH AFFINITY IMMUNOGLOBULIN EPSILON RECEPTOR SUBUNIT ALPHA, IG EPSILON CHAIN C REGION, ... | | Authors: | Davies, A.M, Holdom, M.D, Nettleship, J.E, Beavil, A.J, Owens, R.J, Sutton, B.J. | | Deposit date: | 2011-02-01 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Conformational Changes in Ige Contribute to its Uniquely Slow Dissociation Rate from Receptor Fceri
Nat.Struct.Mol.Biol., 18, 2011
|
|
2C4C
 
 | | Crystal structure of the NADPH-treated monooxygenase domain of MICAL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NEDD9-INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-10-18 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BRY
 
 | | Crystal structure of the native monooxygenase domain of MICAL at 1.45 A resolution | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-05-13 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
8BS8
 
 | | Bovine naive ultralong antibody AbD08 collected at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heavy chain, Light chain | | Authors: | Clarke, J.D, Douangamath, A, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The impact of exchanging the light and heavy chains on the structures of bovine ultralong antibodies.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
2GW8
 
 | | Structure of the PII signal transduction protein of Neisseria meningitidis at 1.85 resolution | | Descriptor: | PII signal transduction protein | | Authors: | Nichols, C.E, Sainsbury, S, Berrow, N.S, Alderton, D, Stammers, D.K, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2006-05-04 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the P(II) signal transduction protein of Neisseria meningitidis at 1.85 A resolution.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
4KQQ
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4KQO
 
 | | Crystal structure of penicillin-binding protein 3 from pseudomonas aeruginosa in complex with piperacillin | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
3G3Z
 
 | | The structure of NMB1585, a MarR family regulator from Neisseria meningitidis | | Descriptor: | Transcriptional regulator, MarR family | | Authors: | Nichols, C.E, Sainsbury, S, Ren, J, Walter, T.S, Verma, A, Stammers, D.K, Saunders, N.J, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-02-03 | | Release date: | 2009-04-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of NMB1585, a MarR-family regulator from Neisseria meningitidis
Acta Crystallogr.,Sect.F, 65, 2009
|
|
