8Q72
 
 | | E. coli plasmid-borne JetABCD(E248A) core in a cleavage-competent state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Circular plasmid DNA (1840-MER), JetA, ... | | Authors: | Roisne-Hamelin, F, Li, Y, Gruber, S. | | Deposit date: | 2023-08-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structural basis for plasmid restriction by SMC JET nuclease.
Mol.Cell, 84, 2024
|
|
6FOE
 
 | | BaxB01 Fab fragment | | Descriptor: | Fab BaxB01 heavy chain, Fab BaxB01 light chain | | Authors: | Hollerweger, J, Schinagl, A, Kerschbaumer, R.J, Scheiflinger, F, Thiele, M, Goettig, P, Brandstetter, H. | | Deposit date: | 2018-02-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Role of the Cysteine 81 Residue of Macrophage Migration Inhibitory Factor as a Molecular Redox Switch.
Biochemistry, 57, 2018
|
|
6S4A
 
 | | Structure of human MTHFD2 in complex with TH9028 | | Descriptor: | (2~{S})-2-[[5-[[2,4-bis(azanyl)-6-oxidanylidene-5~{H}-pyrimidin-5-yl]carbamoylamino]pyridin-2-yl]carbonylamino]-4-(1~{H}-1,2,3,4-tetrazol-5-yl)butanoic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4F
 
 | | Structure of human MTHFD2 in complex with TH9619 | | Descriptor: | (E,4S)-4-[[5-[2-[2,6-bis(azanyl)-4-oxidanylidene-1H-pyrimidin-5-yl]ethanoylamino]-3-fluoranyl-pyridin-2-yl]carbonylamino]pent-2-enedioic acid, ADENOSINE-5'-DIPHOSPHATE, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, ... | | Authors: | Scaletti, E.R, Gustafsson, R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6S4E
 
 | | Structure of human MTHFD2 in complex with TH7299 | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Gustafsson, R, Scaletti, E.R, Bonagas, N, Gustafsson, N.M, Henriksson, M, Abdurakhmanov, E, Andersson, Y, Bengtsson, C, Borhade, S, Desroses, M, Farnegardh, K, Garg, N, Gokturk, C, Haraldsson, M, Iliev, P, Jarvius, M, Jemth, A.S, Kalderen, C, Karsten, S, Klingegard, F, Koolmeister, T, Martens, U, Llona-Minguez, S, Loseva, O, Marttila, P, Michel, M, Moulson, R, Nordstrom, H, Paulin, C, Pham, T, Pudelko, L, Rasti, A, Roos, A.K, Sarno, A, Sandberg, L, Scobie, M, Sjoberg, B, Svensson, R, Unterlass, J.E, Vallin, K, Vo, D, Wiita, E, Warpman-Berglund, U, Homan, E.J, Helleday, T, Stenmark, P. | | Deposit date: | 2019-06-27 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pharmacological targeting of MTHFD2 suppresses acute myeloid leukemia by inducing thymidine depletion and replication stress.
Nat Cancer, 3, 2022
|
|
6QXB
 
 | | NMR structure of peptide 7, characterized by a cis-4-amino-Pro residue, with a significant lower MIC on E. coli | | Descriptor: | PHE-VAL-CAP-TRP-PHE-SER-LYS-PHE-LEU-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Grieco, P, Novellino, E. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-29 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The Outcomes of Decorated Prolines in the Discovery of Antimicrobial Peptides from Temporin-L.
Chemmedchem, 14, 2019
|
|
6QXC
 
 | | NMR structure of peptide 8, characterized by a trans-4-cyclohexyl-Pro, with a dramatic reduction in activity on E. coli ATCC and lost effect on P. aeruginosa. | | Descriptor: | PHE-VAL-TCP-TRP-PHE-SER-LYS-PHE-LEU-GLY-ARG-ILE-LEU-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Merlino, F, Grieco, P, Novellino, E. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-29 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | The Outcomes of Decorated Prolines in the Discovery of Antimicrobial Peptides from Temporin-L.
Chemmedchem, 14, 2019
|
|
3H4V
 
 | | Selective screening and design to identify inhibitors of leishmania major pteridine reductase 1 | | Descriptor: | METHYL 1-(4-{[(2,4-DIAMINOPTERIDIN-6-YL)METHYL]AMINO}BENZOYL)PIPERIDINE-4-CARBOXYLATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1 | | Authors: | Mcluskey, K, Gibellini, F, Hunter, W.N. | | Deposit date: | 2009-04-21 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent pteridine reductase inhibitors to guide antiparasite drug development
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1W0C
 
 | | Inhibition of Leishmania major pteridine reductase (PTR1) by 2,4,6-triaminoquinazoline; structure of the NADP ternary complex. | | Descriptor: | 2,4,6-TRIAMINOQUINAZOLINE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE | | Authors: | Mcluskey, K, Gibellini, F, Carvalho, P, Avery, M, Hunter, W. | | Deposit date: | 2004-06-02 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibition of Leishmania Major Pteridine Reductase by 2,4,6-Triaminoquinazoline: Structure of the Nadph Ternary Complex
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1MJ1
 
 | | FITTING THE TERNARY COMPLEX OF EF-Tu/tRNA/GTP AND RIBOSOMAL PROTEINS INTO A 13 A CRYO-EM MAP OF THE COLI 70S RIBOSOME | | Descriptor: | Elongation Factor Tu, L11 ribosomal protein, Phe-tRNA, ... | | Authors: | Stark, H, Rodnina, M.V, Wieden, H.-J, Zemlin, F, Wintermeyer, W, Vanheel, M. | | Deposit date: | 2002-08-26 | | Release date: | 2002-11-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Ribosome Interactions of Aminoacyl-tRNA and Elongation Factor TU in the
Codon Recognition Complex
Nat.Struct.Biol., 9, 2002
|
|
8S8P
 
 | | Restriction on Ku Inward Translocation Caps Telomere Ends | | Descriptor: | ATP-dependent DNA helicase II subunit 1, ATP-dependent DNA helicase II subunit 2, DNA (5'-D(*AP*CP*AP*CP*AP*CP*AP*CP*AP*CP*CP*CP*AP*CP*AP*CP*AP*CP*CP*AP*C)-3'), ... | | Authors: | Mattarocci, S, Baconnais, S, Roisne-Hamelin, F, Pobiega, S, Alibert, O, Morin, V, Deshayes, A, Veaute, X, Ropars, V, Mazon, G, Busso, D, Fernandez Varela, P, Le Cam, E, Charbonnier, J, Cuniasse, P, Marcand, S. | | Deposit date: | 2024-03-07 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | A Restriction on Ku Inward Translocation Caps Telomere Ends
To Be Published
|
|
8S82
 
 | | Restriction on Ku Inward Translocation Caps Telomere Ends | | Descriptor: | ATP-dependent DNA helicase II subunit 1, ATP-dependent DNA helicase II subunit 2, DNA (5'-D(*AP*CP*AP*CP*AP*CP*AP*CP*AP*CP*CP*CP*AP*CP*AP*CP*AP*CP*CP*AP*C)-3'), ... | | Authors: | Mattarocci, S, Baconnais, S, Roisne-Hamelin, F, Pobiega, S, Alibert, O, Morin, V, Deshayes, A, Veaute, X, Ropars, V, Mazon, G, Busso, D, Fernandez Varela, P, Le Cam, E, Charbonnier, J, Cuniasse, P, Marcand, S. | | Deposit date: | 2024-03-05 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | A Restriction on Ku Inward Translocation Caps Telomere Ends
To Be Published
|
|
8AS8
 
 | | E. coli Wadjet JetABC monomer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, JetA, JetB, ... | | Authors: | Roisne-Hamelin, F, Beckert, B, Li, Y, Myasnikov, A, Gruber, S. | | Deposit date: | 2022-08-18 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA-measuring Wadjet SMC ATPases restrict smaller circular plasmids by DNA cleavage.
Mol.Cell, 82, 2022
|
|
1QIB
 
 | | CRYSTAL STRUCTURE OF GELATINASE A CATALYTIC DOMAIN | | Descriptor: | 72 kDa type IV collagenase, CALCIUM ION, ZINC ION | | Authors: | Dhanaraj, V, Williams, M.G, Ye, Q.-Z, Molina, F, Johnson, L.L, Ortwine, D.F, Pavlovsky, A, Rubin, J.R, Skeean, R.W, White, A.D, Humblet, C, Hupe, D.J, Blundell, T.L. | | Deposit date: | 1999-06-11 | | Release date: | 1999-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of gelatinase A catalytic domain complexed with a hydroxamate inhibitor
Croatica Chemica Acta, 72, 1999
|
|
1QFC
 
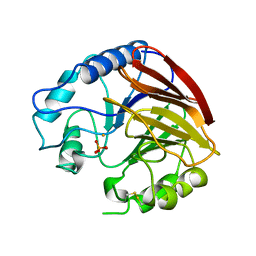 | | STRUCTURE OF RAT PURPLE ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, PHOSPHATE ION, ... | | Authors: | Uppenberg, J, Lindqvist, F, Svensson, C, Ek-Rylander, B, Andersson, G. | | Deposit date: | 1999-04-08 | | Release date: | 2000-04-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a mammalian purple acid phosphatase.
J.Mol.Biol., 290, 1999
|
|
7UCX
 
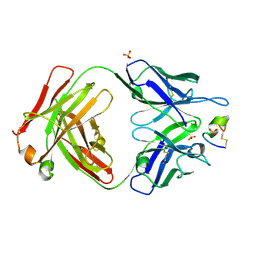 | | LRP8 11H1 Fab complexed to a cyclized CR1 peptide | | Descriptor: | 11H1 Fab Heavy chain, 11H1 Fab Light chain, Cyclized CR1 peptide, ... | | Authors: | Argiriadi, M.A, Deng, K, Egan, D, Gao, L, Gizatullin, F, Harlan, J, Karaoglu, D, Qiu, W, Goodearl, A. | | Deposit date: | 2022-03-17 | | Release date: | 2023-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The use of cyclic peptide antigens to generate LRP8 specific antibodies
Front Drug Discov (Lausanne), 2, 2023
|
|
8S7O
 
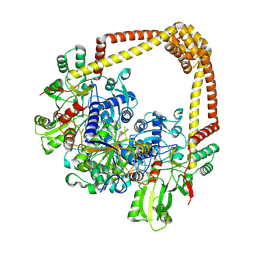 | | M. tuberculosis gyrase holocomplex with 150 bp DNA and BDM71403 | | Descriptor: | 6-[[2-[1-(6-methoxy-1,5-naphthyridin-4-yl)-1,2,3-triazol-4-yl]ethylamino]methyl]-4H-1,4-benzothiazin-3-one, DNA (5'-D(*CP*CP*GP*GP*AP*AP*GP*GP*GP*GP*TP*AP*AP*TP*AP*CP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Gedeon, A, Yab, E, Dinut, A, Sadowski, E, Capton, E, Dreneau, A, Gioia, B, Piveteau, C, Djaout, K, Lecat, E, Wehenkel, A.M, Gubellini, F, Mechaly, A, Alzari, P.M, Deprez, B, Baulard, A, Aubry, A, Willand, N, Petrella, S. | | Deposit date: | 2024-03-04 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | M. tuberculosis gyrase holocomplex with 150 bp DNA and BDM71403
To Be Published
|
|
1BRD
 
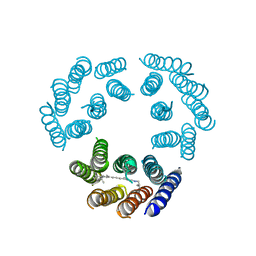 | | Model for the structure of Bacteriorhodopsin based on high-resolution Electron Cryo-microscopy | | Descriptor: | BACTERIORHODOPSIN PRECURSOR, RETINAL | | Authors: | Henderson, R, Baldwin, J.M, Ceska, T.A, Zemlin, F, Beckmann, E, Downing, K.H. | | Deposit date: | 1990-05-23 | | Release date: | 1991-04-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy.
J.Mol.Biol., 213, 1990
|
|
2VVU
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-chloro-N-[(3R)-1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)pyrrolidin-3-yl]thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-11 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VVC
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-chloro-N-[(3S,4S)-1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)-4-methoxypyrrolidin-3-yl]thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-05 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VWL
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID ((3R,5S)-1-{[2-FLUORO-4-(2-OXO-PYRIDIN-1-YL)-PHENYLCARBAMOYL]-METHYL}-5-HYDROXYMETHYL-PYRROLIDIN-3-YL)-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VVV
 
 | | Aminopyrrolidine-related triazole Factor Xa inhibitor | | Descriptor: | 5-chloro-N-[1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)-1H-1,2,4-triazol-3-yl]thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-12 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VWO
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-CHLORO-THIOPHENE-2-CARBOXYLIC ACID ((3S,4S)-4-FLUORO- 1-{[2-FLUORO-4-(2-OXO-2H-PYRIDIN-1-YL)-PHENYLCARBAMOYL]-METHYL}-PYRROLIDIN-3-YL)-AMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VWM
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | (4R)-4-{[(5-chlorothiophen-2-yl)carbonyl]amino}-N-(cyclopropylmethyl)-1-(2-{[2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl]amino}-2-oxoethyl)-L-prolinamide, ACTIVATED FACTOR XA HEAVY CHAIN, FACTOR X LIGHT CHAIN, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
2VWN
 
 | | Aminopyrrolidine Factor Xa inhibitor | | Descriptor: | 5-Chloro-thiophene-2-carboxylic acid ((3S,4S)-1-{[2-fluoro-4-(2-oxo-2H-pyridin-1-yl)-phenylcarbamoyl]-methyl}-4-hydroxy-pyrrolidin-3-yl)-amide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION, ... | | Authors: | Groebke-Zbinden, K, Banner, D.W, Benz, J.M, Blasco, F, Decoret, G, Himber, J, Kuhn, B, Panday, N, Ricklin, F, Risch, P, Schlatter, D, Stahl, M, Unger, R, Haap, W. | | Deposit date: | 2008-06-26 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Design of Novel Aminopyrrolidine Factor Xa Inhibitors from a Screening Hit.
Eur.J.Med.Chem., 44, 2009
|
|
