2POL
 
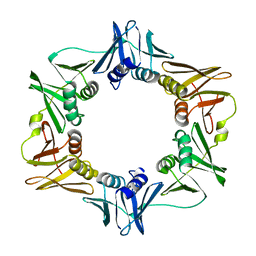 | |
2TPR
 
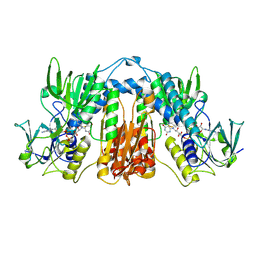 | |
1PLQ
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | MERCURY (II) ION, PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
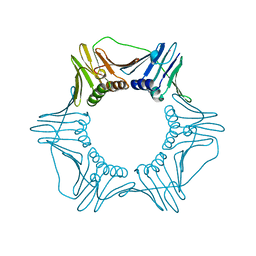
1PLR
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
1EXZ
 
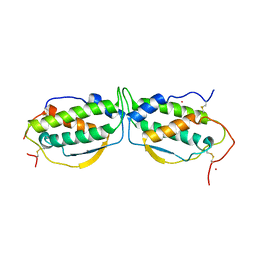 | | STRUCTURE OF STEM CELL FACTOR | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, SAMARIUM (III) ION, ... | | Authors: | Zhang, Z, Zhang, R, Joachimiak, A, Schlessinger, J, Kong, X. | | Deposit date: | 2000-05-05 | | Release date: | 2000-07-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human stem cell factor: implication for stem cell factor receptor dimerization and activation.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
8DGG
 
 | | Structure of glycosylated LAG-3 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silberstein, J.L, Mathews, I.I, Frank, J.A, Chan, K.-W, Fernandez, D, Du, J, Wang, J, Kong, X.-P, Cochran, J.R. | | Deposit date: | 2022-06-23 | | Release date: | 2022-08-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Structural insights reveal interplay between LAG-3 homodimerization, ligand binding, and function.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3GBS
 
 | | Crystal structure of Aspergillus oryzae cutinase | | Descriptor: | Cutinase 1 | | Authors: | Gosser, Y, Lu, Z, Alemu, G, Li, H, Kong, X, Liu, Z, Montclare, J. | | Deposit date: | 2009-02-20 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional studies of Aspergillus oryzae cutinase: enhanced thermostability and hydrolytic activity of synthetic ester and polyester degradation.
J.Am.Chem.Soc., 131, 2009
|
|
3D1F
 
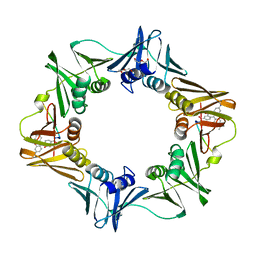 | | Crystal structure of E. coli sliding clamp (beta) bound to a polymerase III peptide | | Descriptor: | 2-[3,6-bis(dimethylamino)xanthen-9-yl]-5-methanoyl-benzoate, DI(HYDROXYETHYL)ETHER, DNA polymerase III subunit beta, ... | | Authors: | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3D1G
 
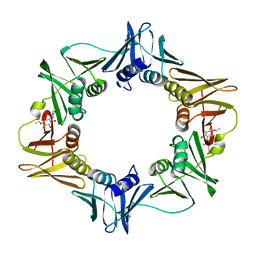 | | Structure of a small molecule inhibitor bound to a DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, [(5R)-5-(2,3-dibromo-5-ethoxy-4-hydroxybenzyl)-4-oxo-2-thioxo-1,3-thiazolidin-3-yl]acetic acid | | Authors: | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3D1E
 
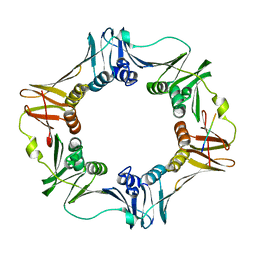 | | Crystal structure of E. coli sliding clamp (beta) bound to a polymerase II peptide | | Descriptor: | DNA polymerase III subunit beta, decamer from polymerase II C-terminal | | Authors: | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4RBR
 
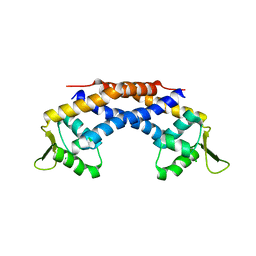 | | Crystal structure of Repressor of Toxin (Rot), a central regulator of Staphylococcus aureus virulence | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator rot | | Authors: | Killikelly, A, Jakoncic, J, Sampson, J.M, Kong, X.-P. | | Deposit date: | 2014-09-12 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Functional Characterization of Repressor of Toxin (Rot), a Central Regulator of Staphylococcus aureus Virulence.
J.Bacteriol., 197, 2015
|
|
6VU2
 
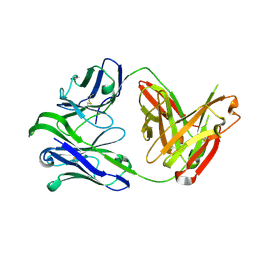 | | M1214_N1 Fab structure | | Descriptor: | M1214 N1 Fab heavy chain, M1214 N1 Fab light chain | | Authors: | Pan, R, Kong, X. | | Deposit date: | 2020-02-14 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA.
Cell Host Microbe, 27, 2020
|
|
3KTZ
 
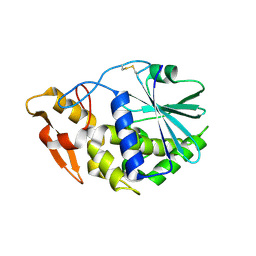 | | Structure of GAP31 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3KU0
 
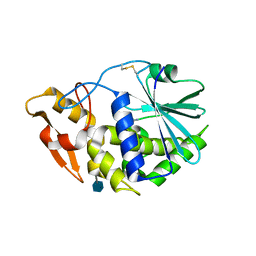 | | Structure of GAP31 with adenine at its binding pocket | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, Ribosome-inactivating protein gelonin | | Authors: | Kong, X.-P. | | Deposit date: | 2009-11-26 | | Release date: | 2010-01-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new activity of anti-HIV and anti-tumor protein GAP31: DNA adenosine glycosidase--structural and modeling insight into its functions.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
3MLS
 
 | |
3MLZ
 
 | | Crystal structure of anti-HIV-1 V3 Fab 3074 in complex with a VI191 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLW
 
 | | Crystal structure of anti-HIV-1 V3 Fab 1006-15D in complex with an MN V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 1006-15D Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 1006-15D Fab light chain, ... | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLT
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a UG1033 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLU
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a ZAM18 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLR
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with a NY5 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLV
 
 | | Crystal structure of anti-HIV-1 V3 Fab 2557 in complex with an NOF V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 2557 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLY
 
 | | Crystal structure of anti-HIV-1 V3 Fab 3074 in complex with a UR29 V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3MLX
 
 | | Crystal structure of anti-HIV-1 V3 Fab 3074 in complex with an MN V3 peptide | | Descriptor: | HIV-1 gp120 third variable region (V3) crown, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab heavy chain, Human monoclonal anti-HIV-1 gp120 V3 antibody 3074 Fab light chain | | Authors: | Kong, X.-P. | | Deposit date: | 2010-04-18 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conserved structural elements in the V3 crown of HIV-1 gp120.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5V6M
 
 | | Crystal Structure of Rabbit Anti-HIV-1 gp120 V3 Fab 10A3 in complex with V3 peptide ConB | | Descriptor: | CALCIUM ION, Envelope glycoprotein gp120 V3 peptide of Con B sequence, Heavy chain of Fab fragment of rabbit anti-HIV1 gp120 V3 mAb 10A3, ... | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Increased epitope complexity correlated with antibody affinity maturation and a novel binding mode revealed by structures of rabbit antibodies against the third variable loop (V3) of HIV-1 gp120.
J. Virol., 2018
|
|
5V6L
 
 | | Crystal Structure of Rabbit Anti-HIV-1 gp120 V3 Fab 10A37 in complex with V3 peptide JR-FL | | Descriptor: | Envelope glycoprotein, v3 region, Heavy chain of Fab fragment of rabbit anti-HIV1 gp120 V3 mAb 10A37, ... | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2017-03-17 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.549 Å) | | Cite: | Increased epitope complexity correlated with antibody affinity maturation and a novel binding mode revealed by structures of rabbit antibodies against the third variable loop (V3) of HIV-1 gp120.
J. Virol., 2018
|
|
