3EIO
 
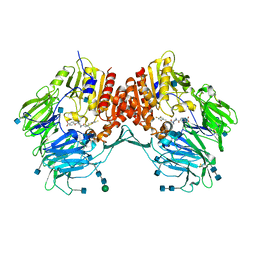 | | Crystal Structure Analysis of DPPIV Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-({4-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl]-1,4-diazepan-1-yl}carbonyl)benzoic acid, ... | | Authors: | Ahn, J.H, Lee, J.-O. | | Deposit date: | 2008-09-17 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and biological evaluation of homopiperazine derivatives with beta-aminoacyl group as dipeptidyl peptidase IV inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3Q8W
 
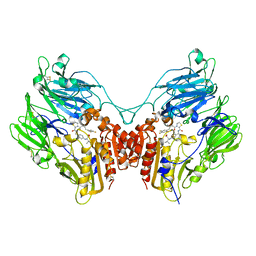 | | A b-aminoacyl containing thiazolidine derivative and DPPIV complex | | Descriptor: | Dipeptidyl peptidase 4, N-(4-{[({(2R)-3-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl]-1,3-thiazolidin-2-yl}carbonyl)amino]methyl}phenyl)-D-valine | | Authors: | Lee, J.O, Song, D.H. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Discovery of b-aminoacyl containing thiazolidine derivatives as potent and selective dipeptidyl peptidase IV inhibitors
To be Published
|
|
7COE
 
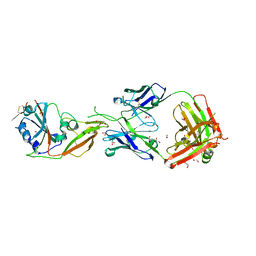 | | Crystal structure of Receptor binding domain of MERS-CoV and KNIH90-F1 Fab complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, ... | | Authors: | Lee, J.Y, Song, J.Y, Lee, H.S, Hong, E, Jang, T.H. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a novel antibody against the spike protein inhibits Middle East respiratory syndrome coronavirus infections.
Sci Rep, 12, 2022
|
|
1AYN
 
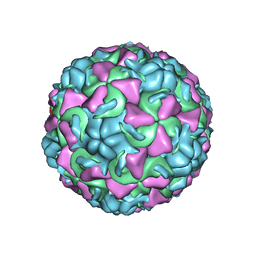 | | HUMAN RHINOVIRUS 16 COAT PROTEIN | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Oliveira, M.A, Zhao, R, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of human rhinovirus 16.
Structure, 1, 1993
|
|
6CX0
 
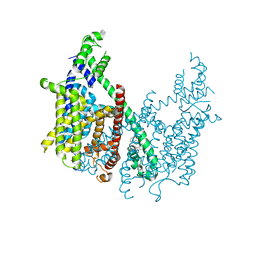 | | Structure of AtTPC1 D376A | | Descriptor: | (1S,3R)-1-(3-{[4-(2-fluorophenyl)piperazin-1-yl]methyl}-4-methoxyphenyl)-2,3,4,9-tetrahydro-1H-beta-carboline-3-carboxylic acid, CALCIUM ION, Two pore calcium channel protein 1 | | Authors: | Kintzer, A.F, Stroud, R.M. | | Deposit date: | 2018-04-02 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1AYM
 
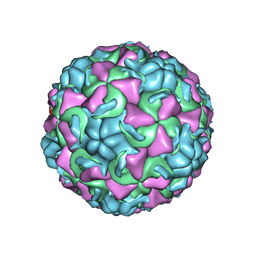 | | HUMAN RHINOVIRUS 16 COAT PROTEIN AT HIGH RESOLUTION | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The refined structure of human rhinovirus 16 at 2.15 A resolution: implications for the viral life cycle.
Structure, 5, 1997
|
|
6E1M
 
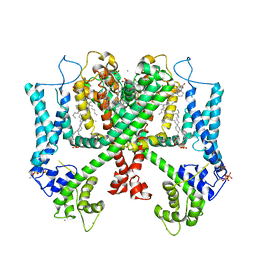 | | Structure of AtTPC1(DDE) reconstituted in saposin A | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CALCIUM ION, PALMITIC ACID, ... | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E1P
 
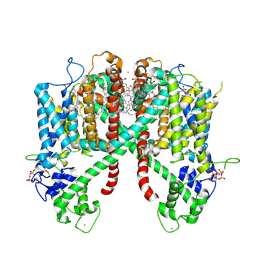 | | Structure of AtTPC1(DDE) in state 2 | | Descriptor: | CALCIUM ION, PALMITIC ACID, Two pore calcium channel protein 1 | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E1N
 
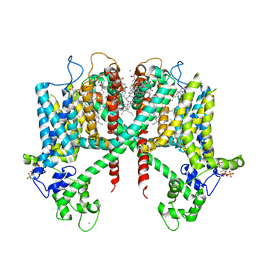 | | Structure of AtTPC1(DDE) in state 1 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, CALCIUM ION, PALMITIC ACID, ... | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6E1K
 
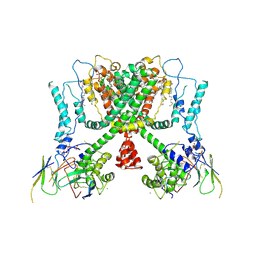 | | Structure of AtTPC1(DDE) reconstituted in saposin A with cat06 Fab | | Descriptor: | CALCIUM ION, PALMITIC ACID, Two pore calcium channel protein 1, ... | | Authors: | Kintzer, A.F, Green, E.M, Cheng, Y, Stroud, R.M. | | Deposit date: | 2018-07-10 | | Release date: | 2018-09-19 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1C8M
 
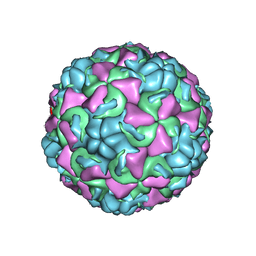 | | REFINED CRYSTAL STRUCTURE OF HUMAN RHINOVIRUS 16 COMPLEXED WITH VP63843 (PLECONARIL), AN ANTI-PICORNAVIRAL DRUG CURRENTLY IN CLINICAL TRIALS | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, HUMAN RHINOVIRUS 16 COAT PROTEIN, ZINC ION | | Authors: | Chakravarty, S, Bator, C.M, Pevear, D.C, Diana, G.D, Rossmann, M.G. | | Deposit date: | 2000-05-26 | | Release date: | 2000-11-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | THE REFINED STRUCTURE OF A PICORNAVIRUS INHIBITOR CURRENTLY IN CLINICAL TRIALS, WHEN COMPLEXED WITH HUMAN RHINOVIRUS 16
to be published
|
|
3GHB
 
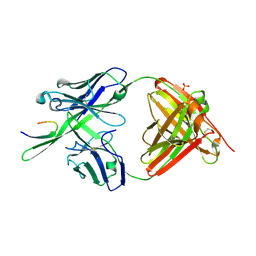 | |
3GHE
 
 | |
3D1E
 
 | | Crystal structure of E. coli sliding clamp (beta) bound to a polymerase II peptide | | Descriptor: | DNA polymerase III subunit beta, decamer from polymerase II C-terminal | | Authors: | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3D1F
 
 | | Crystal structure of E. coli sliding clamp (beta) bound to a polymerase III peptide | | Descriptor: | 2-[3,6-bis(dimethylamino)xanthen-9-yl]-5-methanoyl-benzoate, DI(HYDROXYETHYL)ETHER, DNA polymerase III subunit beta, ... | | Authors: | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3D1G
 
 | | Structure of a small molecule inhibitor bound to a DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, [(5R)-5-(2,3-dibromo-5-ethoxy-4-hydroxybenzyl)-4-oxo-2-thioxo-1,3-thiazolidin-3-yl]acetic acid | | Authors: | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1RHI
 
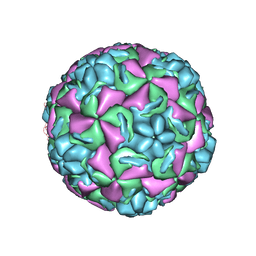 | | HUMAN RHINOVIRUS 3 COAT PROTEIN | | Descriptor: | CALCIUM ION, HUMAN RHINOVIRUS 3 COAT PROTEIN | | Authors: | Zhao, R, Rossmann, M.G. | | Deposit date: | 1996-06-17 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human rhinovirus 3 at 3.0 A resolution.
Structure, 4, 1996
|
|
1QJX
 
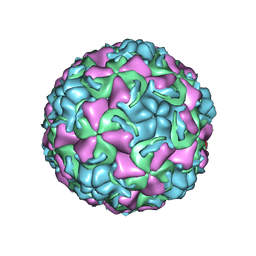 | | HUMAN RHINOVIRUS 16 COAT PROTEIN IN COMPLEX WITH ANTIVIRAL COMPOUND WIN68934 | | Descriptor: | 2,6-DIMETHYL-1-(3-[3-METHYL-5-ISOXAZOLYL]-PROPANYL)-4-[4-METHYL-2H-TETRAZOL-2-YL]-PHENOL, MYRISTIC ACID, PROTEIN VP1, ... | | Authors: | Hadfield, A.T, Diana, G.D, Rossmann, M.G. | | Deposit date: | 1999-07-06 | | Release date: | 1999-07-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Analysis of Three Structurally Related Antiviral Compounds in Complex with Human Rhinovirus 16
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3MCG
 
 | |
2MCG
 
 | |
