3KT2
 
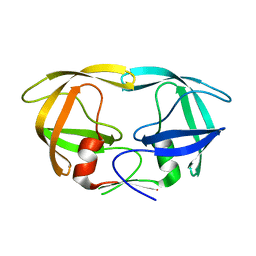 | | Crystal Structure of N88D mutant HIV-1 Protease | | Descriptor: | Protease | | Authors: | Bihani, S.C, Das, A, Prashar, V, Ferrer, J.L, Hosur, M.V. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Resistance mechanism revealed by crystal structures of unliganded nelfinavir-resistant HIV-1 protease non-active site mutants N88D and N88S.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
3N3I
 
 | | Crystal Structure of G48V/C95F tethered HIV-1 Protease/Saquinavir complex | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, Protease | | Authors: | Prashar, V, Bihani, S.C, Das, A, Rao, D.R, Hosur, M.V. | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Insights into the mechanism of drug resistance: X-ray structure analysis of G48V/C95F tethered HIV-1 protease dimer/saquinavir complex
Biochem.Biophys.Res.Commun., 396, 2010
|
|
5I4W
 
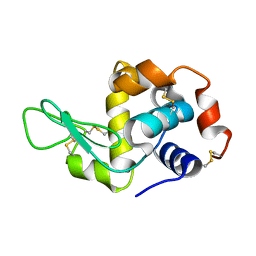 | | Exploring the onset of lysozyme denaturation by urea | | Descriptor: | Lysozyme C | | Authors: | Hosur, M.V, Raskar, T. | | Deposit date: | 2016-02-13 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-dependent X-ray diffraction studies on urea/hen egg white lysozyme complexes reveal structural changes that indicate onset of denaturation
Sci Rep, 6, 2016
|
|
5I54
 
 | |
5I53
 
 | |
5I4X
 
 | |
5I4Y
 
 | |
1NOV
 
 | | NODAMURA VIRUS | | Descriptor: | NODAMURA VIRUS COAT PROTEINS | | Authors: | Natarajan, P, Johnson, J.E. | | Deposit date: | 1997-09-16 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Resolution of space-group ambiguity and structure determination of nodamura virus to 3.3 A resolution from pseudo-R32 (monoclinic) crystals.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1LYB
 
 | | CRYSTAL STRUCTURES OF NATIVE AND INHIBITED FORMS OF HUMAN CATHEPSIN D: IMPLICATIONS FOR LYSOSOMAL TARGETING AND DRUG DESIGN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATHEPSIN D, PEPSTATIN, ... | | Authors: | Baldwin, E.T, Bhat, T.N, Gulnik, S, Erickson, J.W. | | Deposit date: | 1993-04-22 | | Release date: | 1994-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1LYA
 
 | | CRYSTAL STRUCTURES OF NATIVE AND INHIBITED FORMS OF HUMAN CATHEPSIN D: IMPLICATIONS FOR LYSOSOMAL TARGETING AND DRUG DESIGN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATHEPSIN D, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Baldwin, E.T, Bhat, T.N, Gulnik, S, Erickson, J.W. | | Deposit date: | 1993-04-22 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
5XWK
 
 | |
3IX0
 
 | |
1LYW
 
 | | CATHEPSIN D AT PH 7.5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CATHEPSIN D | | Authors: | Lee, A.Y, Gulnik, S.V, Erickson, J.W. | | Deposit date: | 1998-06-30 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational switching in an aspartic proteinase.
Nat.Struct.Biol., 5, 1998
|
|
1HVC
 
 | | CRYSTAL STRUCTURE OF A TETHERED DIMER OF HIV-1 PROTEASE COMPLEXED WITH AN INHIBITOR | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2S,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Bhat, T.N, Baldwin, E.T, Erickson, J.W. | | Deposit date: | 1994-06-22 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a tethered dimer of HIV-1 proteinase complexed with an inhibitor.
Nat.Struct.Biol., 1, 1994
|
|
1HVS
 
 | | STRUCTURAL BASIS OF DRUG RESISTANCE FOR THE V82A MUTANT OF HIV-1 PROTEASE: BACKBONE FLEXIBILITY AND SUBSITE REPACKING | | Descriptor: | HIV-1 PROTEASE, N-{1-BENZYL-(2R,3S)-2,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Baldwin, E.T, Bhat, T.N, Liu, B, Pattabiraman, N, Erickson, J.W. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of drug resistance for the V82A mutant of HIV-1 proteinase.
Nat.Struct.Biol., 2, 1995
|
|
