8XW3
 
 | |
8XVY
 
 | |
8XVZ
 
 | |
8XW4
 
 | | Cryo-EM structure of TMEM63B-Digitonin state | | Descriptor: | CSC1-like protein 2 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XAJ
 
 | | Cryo-EM structure of OSCA1.2-liposome-inside-in open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XVX
 
 | |
8XS0
 
 | |
6L2G
 
 | | Crystal structure of Aspergillus fumigatus mitochondrial acetyl-CoA acetyltransferase | | Descriptor: | Acetyl-CoA-acetyltransferase, putative | | Authors: | Zhang, Y, Wei, W, Raimi, O.G, Ferenbach, A.T, Fang, W. | | Deposit date: | 2019-10-03 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Aspergillus fumigatus Mitochondrial Acetyl Coenzyme A Acetyltransferase as an Antifungal Target.
Appl.Environ.Microbiol., 86, 2020
|
|
6L2C
 
 | | Crystal structure of Aspergillus fumigatus mitochondrial acetyl-CoA acetyltransferase in complex with CoA | | Descriptor: | Acetyl-CoA-acetyltransferase, putative, COENZYME A | | Authors: | Zhang, Y, Wei, W, Raimi, O.G, Ferenbach, A.T, Fang, W. | | Deposit date: | 2019-10-03 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Aspergillus fumigatus Mitochondrial Acetyl Coenzyme A Acetyltransferase as an Antifungal Target.
Appl.Environ.Microbiol., 86, 2020
|
|
8IHM
 
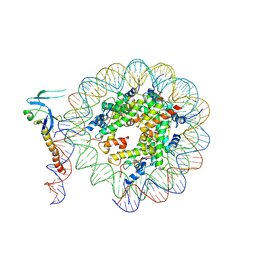 | | Eaf3 CHD domain bound to the nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (164-MER), DNA (165-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHN
 
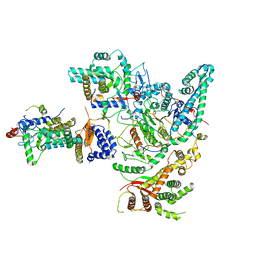 | | Cryo-EM structure of the Rpd3S core complex | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, Histone H3, ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8IHT
 
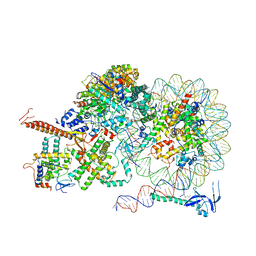 | | Rpd3S bound to the nucleosome | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, DNA (164-MER), ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
7K65
 
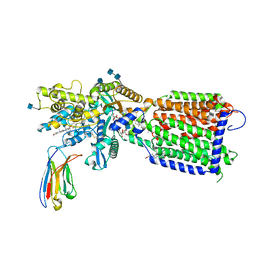 | | Hedgehog receptor Patched (PTCH1) in complex with conformation selective nanobody TI23 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ... | | Authors: | Zhang, Y, Bulkley, D.P, Liang, J, Manglik, A, Cheng, Y, Beachy, P.A. | | Deposit date: | 2020-09-18 | | Release date: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Hedgehog pathway activation through nanobody-mediated conformational blockade of the Patched sterol conduit.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6CND
 
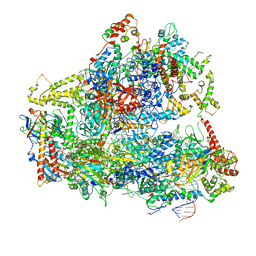 | | Yeast RNA polymerase III natural open complex (nOC) | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CNB
 
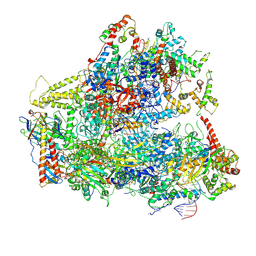 | | Yeast RNA polymerase III initial transcribing complex | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CNC
 
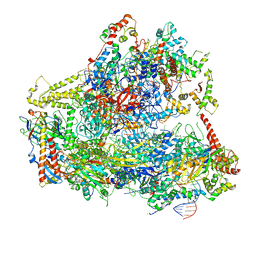 | | Yeast RNA polymerase III open complex | | Descriptor: | DNA (71-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
6CNF
 
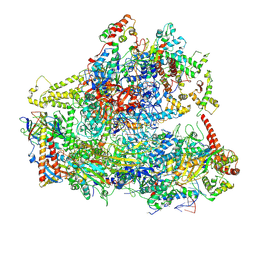 | | Yeast RNA polymerase III elongation complex | | Descriptor: | DNA (79-MER), DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2018-03-08 | | Release date: | 2018-08-22 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural visualization of RNA polymerase III transcription machineries.
Cell Discov, 4, 2018
|
|
5W65
 
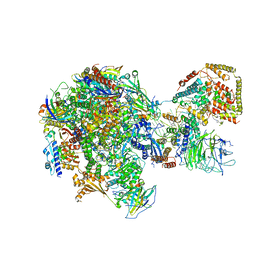 | | RNA polymerase I Initial Transcribing Complex State 2 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-08-02 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W64
 
 | | RNA Polymerase I Initial Transcribing Complex State 1 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-26 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W5Y
 
 | | RNA polymerase I Initial Transcribing Complex | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-26 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
5W66
 
 | | RNA polymerase I Initial Transcribing Complex State 3 | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Han, Y, He, Y. | | Deposit date: | 2017-06-16 | | Release date: | 2017-07-26 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Elife, 6, 2017
|
|
8WNH
 
 | |
8WMT
 
 | |
8WWS
 
 | | Crystal structure of cis-epoxysuccinate hydrolase from Klebsiella oxytoca with L(+)-tartaric acid | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Han, Y, Kong, X.D, Li, J, Xu, J.H. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Insights of a cis -Epoxysuccinate Hydrolase Facilitate the Development of Robust Biocatalysts for the Production of l-(+)-Tartrate.
Biochemistry, 63, 2024
|
|
4G8K
 
 | |
