8FYQ
 
 | | MicroED structure of Proteinase K from argon milled lamellae | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martynowycz, M.W, Shiriaeva, A, Clabbers, M.T.B, Nicolas, W.J, Weaver, S.J, Hattne, J, Gonen, T. | | Deposit date: | 2023-01-26 | | Release date: | 2023-03-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.4 Å) | | Cite: | A robust approach for MicroED sample preparation of lipidic cubic phase embedded membrane protein crystals.
Nat Commun, 14, 2023
|
|
8FYN
 
 | | MicroED structure of A2A from plasma milled lamellae | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Martynowycz, M.W, Shiriaeva, A, Clabbers, M.T.B, Nicolas, W.J, Weaver, S.J, Hattne, J, Gonen, T. | | Deposit date: | 2023-01-26 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (2 Å) | | Cite: | A robust approach for MicroED sample preparation of lipidic cubic phase embedded membrane protein crystals.
Nat Commun, 14, 2023
|
|
8FYS
 
 | | MicroED structure of Proteinase K from nitrogen milled lamellae | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Martynowycz, M.W, Shiriaeva, A, Clabbers, M.T.B, Nicolas, W.J, Weaver, S.J, Hattne, J, Gonen, T. | | Deposit date: | 2023-01-26 | | Release date: | 2023-03-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.8 Å) | | Cite: | A robust approach for MicroED sample preparation of lipidic cubic phase embedded membrane protein crystals.
Nat Commun, 14, 2023
|
|
8FYO
 
 | | MicroED structure of Proteinase K from lamellae milled from multiple plasma sources | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martynowycz, M.W, Shiriaeva, A, Clabbers, M.T.B, Nicolas, W.J, Weaver, S.J, Hattne, J, Gonen, T. | | Deposit date: | 2023-01-26 | | Release date: | 2023-05-24 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.39 Å) | | Cite: | A robust approach for MicroED sample preparation of lipidic cubic phase embedded membrane protein crystals.
Nat Commun, 14, 2023
|
|
4ZNN
 
 | | MicroED structure of the segment, GVVHGVTTVA, from the A53T familial mutant of Parkinson's disease protein, alpha-synuclein residues 47-56 | | Descriptor: | Alpha-synuclein | | Authors: | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2015-05-05 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.41 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
5A3E
 
 | |
5I9S
 
 | | MicroED structure of proteinase K at 1.75 A resolution | | Descriptor: | Proteinase K, SULFATE ION | | Authors: | Hattne, J, Shi, D, de la Cruz, M.J, Reyes, F.E, Gonen, T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-06-08 | | Last modified: | 2023-08-30 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.75 Å) | | Cite: | Modeling truncated pixel values of faint reflections in MicroED images.
J.Appl.Crystallogr., 49, 2016
|
|
6MXF
 
 | | MicroED structure of thiostrepton at 1.9 A resolution | | Descriptor: | Thiostrepton | | Authors: | Jones, C.G, Martynowycz, M.W, Hattne, J, Fulton, T, Stoltz, B.M, Rodriguez, J.A, Nelson, H.M, Gonen, T. | | Deposit date: | 2018-10-30 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.91 Å) | | Cite: | The CryoEM Method MicroED as a Powerful Tool for Small Molecule Structure Determination.
ACS Cent Sci, 4, 2018
|
|
7KGV
 
 | | Crystal structure of sodium-coupled neutral amino acid transporter SLC38A9 in the N-terminal plugged form | | Descriptor: | Monoclonal antibody Fab heavy chain, Monoclonal antibody Fab light chain, Sodium-coupled neutral amino acid transporter 9 | | Authors: | Lei, H, Mu, X, Hattne, J, Gonen, T. | | Deposit date: | 2020-10-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A conformational change in the N terminus of SLC38A9 signals mTORC1 activation.
Structure, 29, 2021
|
|
7KUH
 
 | | MicroED structure of mVDAC | | Descriptor: | Voltage-dependent anion-selective channel protein 1 | | Authors: | Martynowycz, M.W, Khan, F, Hattne, J, Abramson, J, Gonen, T. | | Deposit date: | 2020-11-25 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.12 Å) | | Cite: | MicroED structure of lipid-embedded mammalian mitochondrial voltage-dependent anion channel.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7JSY
 
 | |
6VHC
 
 | |
8D2T
 
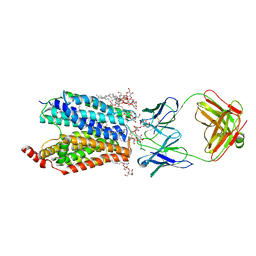 | | Zebrafish MFSD2A isoform B in inward open ligand-free conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2V
 
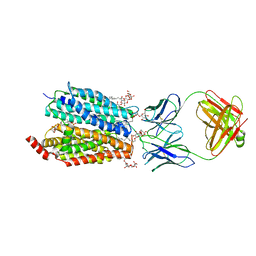 | | Zebrafish MFSD2A isoform B in inward open ligand 1B conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2W
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 2B conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2S
 
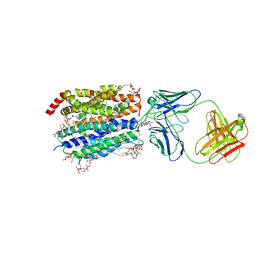 | | Zebrafish MFSD2A isoform B in inward open ligand bound conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2X
 
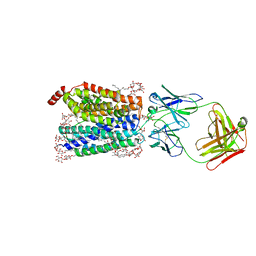 | | Zebrafish MFSD2A isoform B in inward open ligand 3C conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
8D2U
 
 | | Zebrafish MFSD2A isoform B in inward open ligand 1A conformation | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FAB heavy chain, FAB light chain, ... | | Authors: | Nguyen, C, Lei, H.T, Lai, L.T.F, Gallentino, M.J, Mu, X, Matthies, D, Gonen, T. | | Deposit date: | 2022-05-30 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Lipid flipping in the omega-3 fatty-acid transporter.
Nat Commun, 14, 2023
|
|
6VHB
 
 | |
8E52
 
 | | MicroED structure of proteinase K recorded on K2 | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
8E54
 
 | | MicroED structure of triclinic lysozyme recorded on K3 | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
8E53
 
 | | MicroED structure of proteinase K recorded on K3 | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2022-10-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.7 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
8EUN
 
 | | MicroED structure of an Aeropyrum pernix protoglobin metallo-carbene complex | | Descriptor: | Protogloblin ApPgb, benzyl[3,3'-(7,12-diethenyl-3,8,13,17-tetramethylporphyrin-2,18-diyl-kappa~4~N~21~,N~22~,N~23~,N~24~)di(propanoato)(2-)]iron | | Authors: | Danelius, E, Gonen, T, Unge, J.T. | | Deposit date: | 2022-10-19 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.5 Å) | | Cite: | MicroED Structure of a Protoglobin Reactive Carbene Intermediate.
J.Am.Chem.Soc., 145, 2023
|
|
7SW1
 
 | | MicroED structure of proteinase K from a 115 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.85 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SW7
 
 | | MicroED structure of proteinase K from a 530 nm thick lamella measured at 200 kV | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Clabbers, M.T.B, Unge, J, Hattne, J, Gonen, T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.3 Å) | | Cite: | Benchmarking the ideal sample thickness in cryo-EM.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
